Beyond K48 and K63: The Emerging Functions of Atypical Ubiquitin Chains K6, K27, and K29 in Cell Signaling and Disease
This review synthesizes current knowledge on the structures, biological functions, and regulatory mechanisms of the atypical ubiquitin chain linkages K6, K27, and K29.
Beyond K48 and K63: The Emerging Functions of Atypical Ubiquitin Chains K6, K27, and K29 in Cell Signaling and Disease
Abstract
This review synthesizes current knowledge on the structures, biological functions, and regulatory mechanisms of the atypical ubiquitin chain linkages K6, K27, and K29. Once considered rare and enigmatic, these non-canonical chains are now recognized as critical regulators of diverse cellular processes, including innate immunity, mitochondrial quality control, cell cycle progression, and mRNA stability. We explore the specialized tools and methodologies required for their study, address common experimental challenges, and compare their distinct signaling outcomes. Aimed at researchers and drug development professionals, this article highlights the therapeutic potential of targeting these unique ubiquitin codes in human diseases such as cancer, neurodegenerative disorders, and inflammatory conditions.
Decoding the Atypical Trio: Structural and Functional Foundations of K6, K27, and K29 Ubiquitin Chains
Ubiquitination represents one of the most sophisticated post-translational modifications in eukaryotic cells, functioning as a complex molecular code that governs protein fate and function. While the canonical K48 and K63-linked ubiquitin chains have been extensively characterized, recent research has unveiled the critical roles played by atypical linkages—K6, K27, and K29—in regulating specialized cellular processes. These non-canonical chains exhibit unique structural properties and mediate diverse functions beyond protein degradation, including innate immune signaling, transcription regulation, and cell cycle control. This technical review comprehensively examines the assembly mechanisms, structural characteristics, and functional significance of these atypical ubiquitin linkages, with particular emphasis on their implications for therapeutic development. We integrate current methodologies for studying these complex modifications and provide visual schematics of key signaling pathways to elucidate this rapidly evolving field.
The Ubiquitin Code: Fundamental Concepts
Ubiquitin is a small 76-amino acid regulatory protein that is covalently attached to substrate proteins through a sequential enzymatic cascade involving E1 activating, E2 conjugating, and E3 ligase enzymes [1]. The process initiates with E1-mediated ATP-dependent ubiquitin activation, followed by transfer to an E2 enzyme, and culminates in E3-facilitated substrate recognition and ubiquitin transfer [2] [3]. Ubiquitin itself contains seven lysine residues (K6, K11, K27, K29, K33, K48, K63) and an N-terminal methionine (M1) that serve as potential linkage sites for polyubiquitin chain formation [1] [4].
The "ubiquitin code" hypothesis posits that distinct chain topologies—including homotypic, mixed, and branched chains—encode specific functional outcomes that are decoded by ubiquitin-binding domains (UBDs) present in effector proteins [3] [5]. This molecular code exhibits remarkable complexity due to several factors: the diversity of possible linkage combinations, variations in chain length, and the potential for further post-translational modifications on ubiquitin itself [6]. While K48-linked chains predominantly target substrates for proteasomal degradation and K63-linked chains regulate signaling pathways, the functional roles of atypical linkages are more specialized and context-dependent [7].
Table 1: Fundamental Components of the Ubiquitination System
| Component | Number in Humans | Primary Functions |
|---|---|---|
| E1 Enzymes | 2 [4] | Ubiquitin activation via ATP hydrolysis |
| E2 Enzymes | ~40 [2] | Ubiquitin conjugation; influences chain topology |
| E3 Ligases | >600 [2] | Substrate recognition; specific ubiquitin transfer |
| Deubiquitinases (DUBs) | ~100 [4] | Ubiquitin chain removal; signal termination |
Atypical Ubiquitin Linkages: Structural and Functional Diversity
K6-Linked Ubiquitination
K6-linked ubiquitin chains have emerged as crucial regulators in quality control pathways, particularly in mitochondrial homeostasis and the DNA damage response. During mitophagy, the E3 ligase Parkin decorates damaged outer mitochondrial membrane proteins with K6, K11, K48, and K63-linked chains, with K6 and K63 linkages primarily designating mitochondria for autophagic clearance [3]. This process is tightly regulated by deubiquitinating enzymes USP8 and USP30, with the latter showing preference for removing K6-linked chains and thereby antagonizing Parkin-mediated ubiquitination [3].
In the DNA damage response, K6-linked auto-ubiquitination occurs in the BRCA1-BARD1 complex, and K6-linked chains accumulate during replication stress and double-strand break repair [3]. The E3 ligase HUWE1 generates the majority of cellular K6-linked species upon inhibition of valosin-containing protein (VCP/p97/Cdc48), suggesting a role in protein disposal [3]. Beyond degradation-related functions, K6-linked chains also play non-proteolytic roles in innate immunity, where they enhance the DNA-binding capacity of interferon regulatory factor 3 (IRF3) during antiviral responses [3].
K27-Linked Ubiquitination
K27-linked ubiquitination serves as a versatile signaling mechanism in innate immune regulation, with context-dependent outcomes determined by specific E3 ligase-substrate pairs. Multiple TRIM family E3 ligases utilize K27 linkages to modulate antiviral signaling pathways: TRIM23 mediates NEMO ubiquitination leading to NF-κB and IRF3 activation [8], while TRIM26 promotes type I interferon production through interaction with NEMO [8]. Conversely, TRIM40 attenuates antiviral responses by targeting RIG-I and MDA5 for proteasomal degradation [8].
The functional diversity of K27 linkages extends beyond simple activation/inhibition paradigms. For instance, K27-linked ubiquitination of Rhbdd3 recruits the deubiquitinase A20 to remove K63-linked chains from NEMO, thereby preventing excessive NF-κB activation and maintaining signaling homeostasis [8]. Similarly, MARCH8 induces K27-linked ubiquitination of MAVS, leading to its autophagy-mediated degradation and subsequent attenuation of type I interferon production [8]. These examples illustrate how K27 chains can function as scaffolds for protein complex assembly or as degradation signals depending on cellular context.
K29-Linked Ubiquitination
K29-linked ubiquitin chains have traditionally been associated with proteasomal degradation but recent evidence reveals additional roles in transcriptional regulation and cellular stress responses. During the unfolded protein response (UPR), K29-linked ubiquitination of SMC1A and SMC3 proteins within the cohesin complex increases significantly, leading to disrupted formation of transcription initiation complexes and subsequent downregulation of cell proliferation-related genes such as SERTAD1 and NUDT16L1 [9].
Structurally, K29-linked diubiquitin adopts an extended conformation with exposed hydrophobic patches on both ubiquitin moieties, enabling diverse protein interactions [10]. K29 linkages frequently exist within mixed or branched chains containing other linkage types, increasing the combinatorial complexity of the ubiquitin code [10]. The HECT E3 ligase UBE3C collaborates with the deubiquitinase vOTU to assemble and edit K29-linked chains, demonstrating sophisticated regulation of this modification [10].
Table 2: Atypical Ubiquitin Linkages: Functions and Regulatory Enzymes
| Linkage | Key E3 Ligases | Biological Functions | Regulatory DUBs |
|---|---|---|---|
| K6 | Parkin, HUWE1, RNF144A/B | Mitophagy, DNA damage response, antiviral immunity [3] | USP30, USP8 [3] |
| K27 | TRIM23, TRIM26, TRIM40, TRIM21, MARCH8, RNF185, AMFR | Innate immune regulation, NF-κB and IRF3 activation, MAVS and STING regulation [8] | USP13, USP21, USP19 [8] |
| K29 | UBE3C, SKP1-Cullin-Fbx21 | Transcriptional regulation in UPR, proteasomal degradation [9] [8] | vOTU [10] |
| K11 | RNF26, APC/C (with UBE2S) | Cell cycle regulation, STING regulation, innate immunity [8] [3] | USP19 [8] |
| K33 | RNF2 | Suppression of ISG transcription [8] | USP38 [8] |
Branched Ubiquitin Chains: Complexity Multiplied
Branched ubiquitin chains represent a higher level of complexity in the ubiquitin code, where individual ubiquitin monomers are simultaneously modified at multiple sites to create branched architectures. These specialized structures include K11/K48, K29/K48, and K48/K63 branched chains with distinct physiological functions [5]. The synthesis of branched chains frequently involves collaborative efforts between E3 ligases with different linkage specificities. For instance, during NF-κB signaling, TRAF6 and HUWE1 cooperate to produce branched K48/K63 chains, while in yeast, Ufd4 and Ufd2 collaborate to synthesize branched K29/K48 chains on substrates of the ubiquitin fusion degradation pathway [5].
The functional significance of chain branching is particularly evident in processes requiring precise temporal control. During apoptosis, the regulatory protein TXNIP is first modified with non-proteolytic K63-linked chains by ITCH before UBR5 attaches K48 linkages to form branched K48/K63 chains, resulting in proteasomal degradation of TXNIP [5]. This sequential modification represents an efficient mechanism for converting non-degradative signals to degradative marks. Similarly, the anaphase-promoting complex/cyclosome (APC/C) collaborates with UBE2C and UBE2S to form branched K11/K48 chains on mitotic substrates, enhancing their recognition by the proteasome [5].
Diagram 1: Collaborative synthesis of branched ubiquitin chains by E3 ligases with distinct specificities. The sequential addition of different linkage types creates complex architectures with specialized functions, such as enhanced proteasomal recognition.
Methodological Approaches for Studying Atypical Ubiquitination
Affinity Enrichment Strategies
Advanced affinity separation techniques form the cornerstone of modern ubiquitin research, enabling the selective isolation and characterization of ubiquitinated proteins and specific chain types. Immunoaffinity methods utilizing ubiquitin-specific antibodies or linkage-selective antibodies (e.g., for K48, K63, or M1 chains) provide high specificity for targeted applications [6]. The development of K-ε-GG antibodies that recognize the characteristic diglycine remnant left after tryptic digestion of ubiquitinated proteins has revolutionized ubiquitin site identification by mass spectrometry, enabling large-scale mapping of ubiquitination sites [6].
Ubiquitin-binding domains (UBDs) and engineered tandem ubiquitin-binding entities (TUBEs) offer complementary approaches for ubiquitin enrichment. Naturally occurring UBDs, such as the UBA domain of hHR23A (preferring K48 chains) or UIMs of RAP80 (specific for K63 chains), provide inherent linkage selectivity [6]. TUBEs represent significant methodological advances, created by fusing multiple UBDs to generate reagents with avidity effects and enhanced affinity for polyubiquitin chains [6] [7]. Chain-specific TUBEs with nanomolar affinities enable differentiation between ubiquitin linkage types in high-throughput formats, as demonstrated in studies investigating RIPK2 ubiquitination in inflammatory signaling [7].
Mass Spectrometry and Proteomic Approaches
Mass spectrometry-based proteomics has dramatically expanded our understanding of the ubiquitin code. Advanced techniques now allow for system-wide identification of ubiquitination sites, quantification of chain linkage abundance, and even characterization of branched chain architectures. The integration of affinity enrichment with liquid chromatography-tandem mass spectrometry (LC-MS/MS) has enabled the identification of thousands of ubiquitination sites from complex biological samples [6]. Specialized workflows, such as the Ub-ProT method, combine TUBE-based enrichment with protection from trypsinization to analyze native ubiquitin chain length and composition [6].
Table 3: Essential Research Reagents for Studying Atypical Ubiquitin Linkages
| Reagent/Tool | Type | Specific Application | Key Features |
|---|---|---|---|
| K-ε-GG Antibody [6] | Immunoaffinity reagent | Ubiquitination site mapping | Recognizes diglycine remnant after trypsinization |
| UbiSite Antibody [6] | Immunoaffinity reagent | Ubiquitination site mapping | Recognizes ubiquitin C-terminal 13-amino acid peptide |
| Chain-specific TUBEs [7] | Engineered UBD fusion | Linkage-specific ubiquitin enrichment | Nanomolar affinity; selective for K48, K63, etc. |
| Linkage-specific Antibodies [6] | Immunoaffinity reagent | Detection of specific chain types | Antibodies specific for K48, K63, M1 linkages |
| UBE3C-vOTU Complex [10] | Enzymatic system | K29 chain assembly and editing | Enables controlled synthesis of K29-linked chains |
| Dominant-negative Ubiquitin Mutants [7] | Genetic tool | Linkage function analysis | Lysine-to-arginine mutations to block specific linkages |
Experimental Protocols for Key Applications
Protocol: Analysis of Linkage-Specific Ubiquitination Using TUBEs
This protocol enables the capture and assessment of endogenous protein ubiquitination with linkage specificity, particularly useful for evaluating PROTAC efficacy or inflammatory signaling.
Cell Lysis and Sample Preparation
- Culture THP-1 cells under appropriate conditions and treat with stimuli (e.g., 200-500 ng/ml L18-MDP for K63 ubiquitination or PROTAC for K48 ubiquitination) for 30-60 minutes [7].
- Lyse cells using a specialized lysis buffer optimized to preserve polyubiquitination (e.g., containing 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 10% glycerol, plus protease and deubiquitinase inhibitors) [7].
- Clarify lysates by centrifugation at 14,000 × g for 15 minutes at 4°C.
TUBE-Based Affinity Capture
- Coat 96-well plates with chain-selective TUBEs (K48-TUBE, K63-TUBE, or pan-TUBE) according to manufacturer's specifications [7].
- Incubate 100-500 μg of cell lysate with TUBE-coated plates for 2 hours at 4°C with gentle agitation.
- Wash plates 3-5 times with wash buffer (e.g., 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1% NP-40) to remove non-specifically bound proteins.
Detection and Analysis
- Elute bound proteins using 2× Laemmli buffer containing DTT or directly proceed to in-well detection.
- Analyze captured ubiquitinated proteins by immunoblotting with target-specific antibodies (e.g., anti-RIPK2 for inflammatory signaling studies) [7].
- Quantify band intensity using densitometry software to compare linkage-specific ubiquitination across conditions.
Protocol: Assessment of Branched Ubiquitin Chains
This methodology enables the detection and characterization of branched ubiquitin chains, which often contain atypical linkages.
Enrichment of Ubiquitinated Proteins
- Express epitope-tagged ubiquitin (e.g., His-FLAG-ubiquitin) in cells of interest and treat with relevant stimuli or inhibitors.
- Lyse cells under denaturing conditions (e.g., 6 M guanidine-HCl) to preserve ubiquitination states and prevent deubiquitination.
- Perform immobilized metal affinity chromatography (IMAC) under denaturing conditions to purify ubiquitinated proteins.
Chain Topology Analysis
- Digest enriched ubiquitinated proteins with specific proteases (e.g., trypsin, Lys-C) that generate characteristic ubiquitin fragments.
- Analyze digestion products by LC-MS/MS with methods optimized for ubiquitin branch point detection.
- Use specialized software tools to identify diagnostic ions indicative of branched ubiquitin chains.
Validation Experiments
- Express ubiquitin mutants (e.g., K48R, K63R) to perturb specific branching patterns and confirm findings.
- Utilize E3 ligase knockout/knockdown approaches to identify enzymes responsible for branched chain synthesis.
- Implement in vitro reconstitution assays with purified E2 and E3 enzymes to validate branching mechanisms.
Diagram 2: Workflow for linkage-specific ubiquitination analysis. TUBE-based affinity enrichment followed by mass spectrometry or immunoblotting enables precise characterization of ubiquitin chain types in response to cellular stimuli.
Therapeutic Targeting and Future Perspectives
The intricate regulation of cellular processes by atypical ubiquitin linkages presents compelling opportunities for therapeutic intervention. Several strategies are emerging to target these pathways, particularly in oncology and inflammatory diseases. PROTACs (Proteolysis Targeting Chimeras) represent a groundbreaking approach that hijacks E3 ubiquitin ligases to induce targeted protein degradation [7]. These heterobifunctional molecules simultaneously bind to a target protein and an E3 ligase, facilitating ubiquitination and subsequent proteasomal degradation [2]. While current PROTACs primarily exploit a limited set of E3 ligases (e.g., CRBN, VHL), the expanding knowledge of atypical linkage-specific E3s may enable next-generation degraders with improved selectivity [7].
In inflammatory diseases, targeting K63-linked ubiquitination components (TRAF6, Ubc13, Mms2) has shown promise in preclinical models of rheumatoid arthritis and colitis [7]. Similarly, modulation of linear ubiquitination via LUBAC components or the deubiquitinases OTULIN and CYLD offers therapeutic potential for autoinflammatory conditions [4]. The development of small-molecule inhibitors targeting specific E1, E2, and E3 enzymes continues to advance, with several candidates in clinical development for hematological malignancies and solid tumors [2].
Future research directions will likely focus on deciphering the complex crosstalk between different ubiquitin linkage types, understanding the spatial and temporal regulation of chain assembly and disassembly, and developing more sophisticated tools to manipulate specific aspects of the ubiquitin code. The clinical translation of ubiquitin-based therapeutics will require enhanced selectivity to minimize off-target effects and comprehensive biomarker strategies to identify responsive patient populations.
The expanding universe of atypical ubiquitin linkages—K6, K27, K29, and beyond—has fundamentally transformed our understanding of cellular signaling networks. These non-canonical modifications mediate sophisticated regulatory functions beyond protein degradation, including immune response coordination, transcriptional control, and stress adaptation. The structural diversity of ubiquitin chains is further multiplied through branching and mixed linkage formation, creating an exceptionally complex coding system that integrates multiple signals to determine functional outcomes.
Methodological advances in affinity enrichment, mass spectrometry, and chemical biology continue to unravel the intricacies of the ubiquitin code, revealing new biological insights and therapeutic opportunities. As our toolbox for studying and manipulating these modifications expands, so too does our potential to develop novel therapeutic strategies for cancer, inflammatory diseases, neurodegenerative disorders, and infectious diseases. The continued exploration of atypical ubiquitin linkages promises to yield fundamental discoveries about cellular regulation and unlock new avenues for precision medicine.
Ubiquitination is a crucial post-translational modification that regulates a vast array of cellular processes, with the specificity of signaling outcomes largely determined by the topology of ubiquitin chain linkages. Among the eight possible linkage types, K6-linked ubiquitin chains belong to the category of "atypical" chains that are less abundant but increasingly recognized for their specialized functions. These chains are formed when the C-terminus of one ubiquitin molecule creates an isopeptide bond with the lysine at position 6 (K6) of another ubiquitin. Once considered poorly characterized, K6-linked ubiquitination has emerged as a critical regulator in essential quality control pathways, particularly in DNA damage response and mitochondrial homeostasis. The significance of K6 linkages is further highlighted by their implication in neurodegenerative diseases and cancer, positioning them as potential therapeutic targets in the broader landscape of ubiquitin research.
The study of K6-linked chains has historically been constrained by technical challenges, including the scarcity of linkage-specific detection reagents and enzymatic tools for their manipulation. However, recent advances in chemical biology and protein engineering have begun to illuminate the unique properties and functions of these chains. This technical guide synthesizes current understanding of K6-linked ubiquitin chains, with particular emphasis on their mechanistic roles in cellular stress response pathways, detailed experimental approaches for their investigation, and emerging opportunities for therapeutic intervention targeting the ubiquitin system.
Molecular Mechanisms and Functional Roles
K6-Linked Ubiquitination in DNA Damage Response
The involvement of K6-linked ubiquitin chains in maintaining genomic integrity represents a significant aspect of their functional repertoire, with emerging evidence connecting them to DNA repair processes:
BRCA1-BARD1 Association: Early investigations identified K6-linked ubiquitination in the context of the BRCA1-BARD1 complex, a heterodimeric E3 ubiquitin ligase with established tumor suppressor functions in breast and ovarian cancers. This complex has been shown to assemble K6-linked chains both on itself and on associated substrates, suggesting a potential role in DNA repair pathways that is distinct from the canonical functions of BRCA1 in homologous recombination [11] [12]. The precise mechanistic contribution of K6 linkages to DNA damage signaling remains an active area of investigation, but their presence on this critical tumor suppressor complex underscores their potential significance in maintaining genomic stability.
Cellular Stress Response: Beyond specific repair complexes, K6-linked chains demonstrate a broader involvement in cellular stress adaptation. Research has documented that levels of K6- and K33-linked chains increase following DNA damage induced by genotoxic agents, indicating a potential role in the cellular response to genomic insult [11]. This elevation suggests that K6 linkages may participate in signaling networks that detect, process, or resolve DNA lesions, possibly through the regulation of protein recruitment, activity, or stability at damage sites.
Central Role in Mitophagy and Mitochondrial Quality Control
The most extensively characterized function of K6-linked ubiquitin chains lies in the regulation of mitochondrial quality control, particularly in the PINK1-Parkin mediated mitophagy pathway that ensures the selective removal of damaged mitochondria:
Parkin-Mediated Mitophagy: Parkin, an RBR (RING-Between-RING) E3 ubiquitin ligase mutated in familial forms of Parkinson's disease, plays a central role in marking damaged mitochondria for autophagic clearance. Upon mitochondrial depolarization, Parkin translocates to mitochondria and ubiquitinates numerous outer mitochondrial membrane proteins. Research has demonstrated that Parkin assembles K6-linked ubiquitin chains as part of this process, which are important for the efficient recruitment of Parkin to depolarized mitochondria and subsequent mitophagy [13] [11]. The deposition of K6 linkages represents one of several ubiquitin chain types generated by Parkin, creating a complex ubiquitin code that orchestrates the mitophagy cascade.
USP8 Regulation: The deubiquitinating enzyme USP8 (ubiquitin-specific protease 8) has been identified as a key regulator of Parkin-mediated mitophagy through its specific action on K6-linked ubiquitin chains. USP8 preferentially removes K6-linked ubiquitin conjugates from Parkin itself, a process required for the efficient recruitment of Parkin to depolarized mitochondria and their subsequent elimination [13]. This regulatory mechanism represents a critical control point in mitophagy, wherein USP8-mediated deubiquitination of K6-linked chains from Parkin facilitates Parkin activation and translocation. The antagonistic relationship between Parkin (K6 chain assembly) and USP8 (K6 chain disassembly) fine-tunes the mitophagy response, ensuring that mitochondrial clearance occurs only when appropriate.
HUWE1 and Mitofusin-2 Regulation: Beyond the PINK1-Parkin axis, the HECT E3 ligase HUWE1 has been identified as another significant source of K6-linked ubiquitin chains in cells. Pull-down experiments using K6-specific affimers, combined with mass spectrometry analysis, revealed HUWE1 as a major E3 ligase responsible for cellular K6 chains [11]. Specifically, HUWE1 decorates the mitochondrial protein mitofusin-2 (Mfn2) with K6-linked polyubiquitin, positioning it as a regulator of mitochondrial dynamics. Cells lacking HUWE1 or subjected to HUWE1 knockdown show significantly reduced levels of K6 chains, underscoring the importance of this ligase in the K6-linked ubiquitin landscape [11].
Table 1: Key E3 Ligases and DUBs Regulating K6-Linked Ubiquitination in Mitochondrial Quality Control
| Enzyme | Type | Function on K6 Chains | Biological Role |
|---|---|---|---|
| Parkin | RBR E3 Ligase | Assembles K6-linked chains on mitochondrial substrates | Promotes mitophagy; mutations cause Parkinson's disease |
| HUWE1 | HECT E3 Ligase | Major cellular source of K6 chains; modifies Mfn2 | Regulates mitochondrial dynamics; potential tumor suppressor |
| RNF144A/B | RBR E3 Ligase | Assembles K6-, K11-, and K48-linked chains in vitro | DNA damage response; p53 regulation |
| USP8 | Deubiquitinase | Preferentially removes K6-linked chains from Parkin | Facilitates Parkin recruitment to mitochondria; regulates mitophagy efficiency |
| USP30 | Deubiquitinase | Antagonizes Parkin-mediated mitophagy; K6-selective | Mitochondrial DUB; counteracts mitophagy on mitochondrial surface |
Branched Ubiquitin Chains Involving K6 Linkages
Beyond homogeneous K6-linked chains, recent research has revealed that K6 linkages can form part of more complex branched ubiquitin architectures, significantly expanding their signaling potential:
K6/K48-Branched Chains: Parkin has been demonstrated to synthesize branched ubiquitin chains containing both K6 and K48 linkages, creating a hybrid degradation signal that may combine features of both linkage types [12]. These branched architectures potentially enable more sophisticated regulation of substrate fate than homogeneous chains, possibly integrating degradative signals with specialized regulatory functions.
Other K6-Containing Branches: Evidence also exists for the formation of K6/K11-branched chains, although the physiological functions of these specific architectures remain less defined compared to K6/K48-branched chains [12]. The formation of such diverse branched structures highlights the complexity of the ubiquitin code and suggests that K6 linkages may serve specialized functions when positioned at branch points within polyubiquitin chains.
Quantitative Profiling of K6-Linked Ubiquitination
The investigation of K6-linked ubiquitination relies on quantitative assessments of its abundance, dynamics, and enzymatic regulation under various physiological conditions. The following table summarizes key quantitative findings from recent research:
Table 2: Quantitative Data on K6-Linked Ubiquitin Chain Properties and Functions
| Parameter | Value/Measurement | Experimental Context | Significance |
|---|---|---|---|
| Relative Abundance | Increased after DNA damage | Cellular response to genotoxic stress | Suggests role in DNA damage response [11] |
| DUB Resistance | Resists cleavage by most deubiquitinases | Compared to K27-Ub2 which resists USP2, USP5, Ubp6 | K6-specific DUBs required for regulation [14] |
| HUWE1 Dependency | Significantly reduced K6 levels in HUWE1-/- cells | Pull-downs with K6-specific affimers | HUWE1 is major source of cellular K6 chains [11] |
| Affimer Binding | n = 0.46 (2:1 affimer:diUb complex) | ITC measurements with K6 diUb | Dimerized affimer recognizes K6 linkage specifically [11] |
| USP8 Effect | Delayed but not abolished Parkin recruitment | siRNA knockdown of USP8 | USP8 critical for timely mitophagy initiation [13] |
| Branched Chain Formation | Parkin synthesizes K6/K48-branched chains | In vitro ubiquitination assays | Expands signaling capacity beyond homotypic chains [12] |
Experimental Protocols and Methodologies
siRNA Screening for DUBs Regulating Mitophagy
The identification of USP8 as a regulator of Parkin-mediated mitophagy through its action on K6-linked chains was achieved through a comprehensive siRNA screening approach:
Library Design: The screen targeted 87 putative deubiquitinating enzymes (DUBs) encoded by the human genome using a pre-designed siRNA library [13]. This comprehensive coverage ensured that both known and potentially novel regulators of mitophagy could be identified.
Cell System: Screening was performed in U2OS cells stably expressing GFP-Parkin, providing a consistent expression system for monitoring Parkin translocation. The use of GFP-tagged Parkin enabled quantitative tracking of its movement from cytosol to mitochondria following induction of mitochondrial damage [13].
Induction and Assessment: Mitochondrial depolarization was induced using carbonyl cyanide m-chlorophenyl hydrazone (CCCP), a protonophore that dissipates mitochondrial membrane potential. Parkin recruitment was assessed after 1 hour of CCCP treatment by fluorescence microscopy, with USP8 knockdown identified as the only condition that significantly impaired Parkin translocation at this time point [13].
Validation Experiments: Secondary validation included time-lapse microscopy to precisely quantify the kinetics of Parkin recruitment, immunoblotting to confirm USP8 protein reduction, and rescue experiments with FLAG-USP8 expression plasmids to establish specificity [13]. Mitophagy efficiency was further assessed after 24 hours of CCCP treatment by monitoring the loss of mitochondrial markers like TOM20.
Development and Application of K6-Linkage Specific Affimers
The generation of high-affinity, linkage-specific binding reagents has been instrumental in advancing the study of K6-linked ubiquitination:
Affimer Technology: Affimers are small (12-kDa) non-antibody scaffolds based on the cystatin fold, featuring randomized surface loops that enable selection of high-affinity binders from large libraries (~10^10 variants) [11]. Selections were performed against K6-linked diUb to isolate specific binders.
Specificity Characterization: The resulting K6 affimer was characterized using isothermal titration calorimetry (ITC), showing tight binding to K6-diUb (n = 0.46, indicating 2:1 affimer:diUb complex formation) with no detectable binding to K33-diUb [11]. Surface plasmon resonance (SPR) further confirmed linkage specificity through very slow off-rates specifically for K6-linked chains.
Application in Western Blotting: Site-specifically biotinylated K6 affimers enabled detection of K6-diUb with high linkage specificity in western blotting applications, showing only minimal cross-reactivity with other chain types [11]. This represented a significant advancement as linkage-specific antibodies for K6 chains were previously unavailable.
Structural Basis of Specificity: X-ray crystal structures of K6 affimer bound to K6-diUb revealed the mechanistic basis for linkage specificity, showing that the affimer dimerizes to create two binding sites that engage the I44 patches of both ubiquitin moieties in a K6-diUb molecule with defined spacing and orientation [11]. This mode of recognition mimics naturally occurring ubiquitin-binding domains that achieve specificity through avidity effects.
Biochemical Reconstitution of K6-Linked Ubiquitination
In vitro reconstitution of K6-linked ubiquitination provides a controlled system for mechanistic studies:
Enzyme Selection: Parkin and HUWE1 serve as the primary E3 ligases for generating K6 linkages. Parkin requires activation through phosphorylation by PINK1 and interaction with phospho-ubiquitin, while HUWE1 functions independently of these modifications [11] [12].
Reaction Conditions: Standard ubiquitination reactions include E1 activating enzyme (UBA1), specific E2 conjugating enzymes, the E3 ligase of interest, ubiquitin, and ATP in appropriate buffer. For Parkin, pre-activation with PINK1 or inclusion of phospho-ubiquitin is essential [13] [12].
Chain Analysis: Reaction products are typically analyzed by western blotting with linkage-specific reagents (affimers or antibodies), mass spectrometry for linkage identification, and deubiquitinase treatment with linkage-specific DUBs to confirm chain topology [11].
Pathway Visualization and Signaling Networks
The integration of K6-linked ubiquitination into cellular signaling pathways, particularly in mitochondrial quality control, involves complex regulatory networks that can be visualized through the following diagram:
Diagram 1: K6-Linked Ubiquitination in the PINK1-Parkin Mitophagy Pathway. This diagram illustrates the sequence of events from mitochondrial damage to mitophagy initiation, highlighting the role of K6-linked ubiquitination and its regulation by USP8.
The experimental workflow for investigating K6-linked ubiquitination combines biochemical, cellular, and analytical approaches, as visualized in the following methodology diagram:
Diagram 2: Experimental Workflow for Investigating K6-Linked Ubiquitination. This diagram outlines the key methodological approaches used to identify and characterize the functions of K6-linked ubiquitin chains.
Research Reagent Solutions Toolkit
The investigation of K6-linked ubiquitin chains requires specialized reagents and tools designed to address the unique challenges of studying this atypical ubiquitin linkage. The following table compiles essential research reagents for experimental work in this field:
Table 3: Essential Research Reagents for Studying K6-Linked Ubiquitination
| Reagent/Tool | Type | Specific Function | Key Applications |
|---|---|---|---|
| K6-Linkage Specific Affimers | Engineered binding proteins | Recognize K6-linked diUb with high specificity | Western blotting, immunofluorescence, pull-down assays [11] |
| Recombinant Parkin | RBR E3 ligase | Assembles K6-linked and K6/K48-branched chains in vitro | Biochemical reconstitution of ubiquitination [13] [12] |
| Recombinant HUWE1 | HECT E3 ligase | Major cellular source of K6 linkages | In vitro ubiquitination, identification of K6 substrates [11] |
| USP8 Reagents | Deubiquitinating enzyme | Preferentially removes K6-linked chains from Parkin | Regulation studies, mitophagy modulation [13] |
| K6-Ub2 Chemical Standards | Chemically synthesized diUb | Defined K6-linked ubiquitin dimers | Method calibration, binding studies, DUB characterization [14] |
| Linkage-Specific DUBs | Deubiquitinating enzymes | Cleave specific ubiquitin linkages | Chain topology verification, substrate validation [14] |
| siRNA Libraries | Gene silencing reagents | Targeted knockdown of ubiquitin system components | Functional screens for pathway regulators [13] |
The study of K6-linked ubiquitin chains has evolved from initial observations to mechanistic understanding of their specialized functions in critical cellular quality control pathways. As detailed in this technical guide, K6 linkages play non-redundant roles in Parkin-mediated mitophagy and DNA damage response, operating through specific E3 ligases and regulated by dedicated deubiquitinating enzymes like USP8. The ongoing development of sophisticated research tools, particularly linkage-specific affimers and chemical biology approaches for branched chain synthesis, continues to accelerate discovery in this field.
Looking forward, several key challenges and opportunities merit attention. First, the physiological contexts that specifically trigger K6-linked ubiquitination, as opposed to other linkage types, require further elucidation. Second, the structural basis for recognition of K6-linked chains by downstream effectors remains largely unexplored, representing a significant knowledge gap. Third, the therapeutic potential of modulating K6-linked signaling in diseases such as Parkinson's and cancer warrants systematic investigation, particularly through the development of small molecule inhibitors or activators of relevant E3 ligases and DUBs. As these research directions advance, K6-linked ubiquitin chains will undoubtedly continue to reveal new insights into the complexity of ubiquitin signaling and its manipulation for therapeutic benefit.
Ubiquitination is a crucial post-translational modification that regulates nearly all aspects of cellular function through the covalent attachment of ubiquitin to target proteins. While the roles of K48- and K63-linked polyubiquitin chains in proteasomal degradation and signal transduction, respectively, are well-established, the functions of atypical ubiquitin chains (K6, K11, K27, K29, and K33) represent a frontier in ubiquitin research [8] [15]. Among these, K27-linked ubiquitin chains have emerged as critical regulators of antiviral innate immune signaling and inflammatory pathways, distinguished by their unique structural and biochemical properties [14]. K27-linked chains exhibit remarkable resistance to deubiquitinating enzymes (DUBs) and adopt distinct conformational states that enable specific interactions with downstream signaling components [14] [16]. This comprehensive review examines the multifaceted roles of K27-linked ubiquitination in innate immunity, detailing the molecular mechanisms, regulatory networks, and experimental approaches that define this rapidly advancing field within the broader context of atypical ubiquitin chain biology.
Structural and Functional Uniqueness of K27-Linked Ubiquitin Chains
Distinct Biochemical and Structural Properties
K27-linked ubiquitin chains possess unique characteristics that differentiate them from other ubiquitin linkage types. Structural analyses using NMR spectroscopy and small-angle neutron scattering reveal that K27-linked di-ubiquitin (K27-Ub2) adopts predominantly open conformations in solution, with minimal non-covalent interdomain contacts between ubiquitin units [14]. This structural arrangement stands in contrast to the more compact conformations observed in K48-linked chains. The proximal ubiquitin unit in K27-Ub2 exhibits the largest chemical shift perturbations among all ubiquitin linkage types, indicating significant structural reorganization upon chain formation [14].
A defining feature of K27-linked ubiquitin is its resistance to deubiquitination by most deubiquitinating enzymes. Screening against multiple DUB families including USP2, USP5 (IsoT), and Ubp6 demonstrated that K27-Ub2 resists cleavage, whereas other linkages are efficiently processed [14] [16]. This exceptional stability likely contributes to the sustained signaling functions of K27-linked ubiquitination in immune pathways and suggests specialized regulatory mechanisms for its reversal in cellular contexts.
Recognition by Ubiquitin-Binding Domains
Despite its unique structure, K27-linked ubiquitin demonstrates unexpected versatility in receptor recognition. Structural data indicate that K27-Ub2 can be specifically recognized by the UBA2 domain of the proteasomal shuttle protein hHR23a through bidentate interactions similar to those observed with K48-Ub2 [16]. This binding specificity suggests that K27-linked chains may interface with protein quality control systems while maintaining distinct signaling functions in immune regulation.
Table 1: Key Structural and Biochemical Properties of K27-Linked Ubiquitin Chains
| Property | Characteristic | Functional Implication |
|---|---|---|
| Solution Conformation | Open conformations with minimal non-covalent interdomain contacts | Enables bidentate binding to specific receptors |
| DUB Resistance | Resistant to cleavage by most deubiquitinases (USP2, USP5, Ubp6) | Provides signaling stability and persistence |
| Structural Perturbation | Largest CSPs in proximal Ub among all linkages | Significant structural reorganization upon chain formation |
| Receptor Binding | Binds UBA2 domain of hHR23a similarly to K48-Ub2 | Potential interface with protein quality control systems |
| Chain Architecture | Homogeneous chains, potential for branched formations | Creates diverse recognition platforms |
Molecular Mechanisms of K27-Linked Ubiquitination in Antiviral Signaling
Regulation of Nucleic Acid Sensing Pathways
K27-linked ubiquitination serves as a pivotal regulatory mechanism in multiple innate immune signaling cascades triggered by viral nucleic acid detection. The RIG-I/MAVS pathway, which senses cytoplasmic viral RNA, is extensively modulated by K27-linked ubiquitination through various E3 ligases [8] [17]. TRIM21 catalyzes K27-linked ubiquitination of MAVS, enhancing type I interferon production, while MARCH8 mediates K27-linked ubiquitination of MAVS that induces its autophagy-mediated degradation, thereby restricting the type I interferon response [8]. This opposing regulation illustrates how different E3 ligases utilizing the same linkage type can produce divergent functional outcomes in antiviral signaling.
In the cGAS-STING pathway responsible for cytoplasmic DNA sensing, multiple E3 ligases employ K27-linked ubiquitination to regulate signaling activity. RNF185 mediates K27-linked ubiquitination of cGAS, promoting IRF3 activation and subsequent type I interferon production [8]. Similarly, the endoplasmic reticulum-resident E3 ligase AMFR (gp78) catalyzes K27-linked ubiquitination of STING, facilitating TBK1 recruitment and IRF3 activation [8] [18]. These modifications highlight the critical positioning of K27-linked ubiquitination at key signaling hubs to coordinate appropriate immune responses to diverse viral pathogens.
Modulation of Transcriptional Activation and Inflammatory Responses
K27-linked ubiquitination directly regulates key transcriptional factors and signaling complexes that control interferon and inflammatory cytokine production. TRIM23 catalyzes K27-linked ubiquitination of NEMO (IKKγ), leading to activation of both NF-κB and IRF3 pathways [8]. Additionally, K27-linked ubiquitination of NEMO by other E3 ligases recruits the deubiquitinase A20 to remove K63-linked chains from NEMO, preventing excessive NF-κB activation and illustrating the cross-regulatory potential between different ubiquitin linkage types [8].
Table 2: E3 Ligases and DUBs Regulating K27-Linked Ubiquitination in Innate Immunity
| Enzyme | Substrate | Functional Outcome | Regulatory Effect |
|---|---|---|---|
| TRIM23 | NEMO | Leads to NF-κB and IRF3 activation | Positive regulation |
| TRIM26 | NEMO | Increases type I IFN and cytokine production | Positive regulation |
| TRIM21 | MAVS | Enhances type I IFN production | Positive regulation |
| MARCH8 | MAVS | Induces autophagy-mediated degradation | Negative regulation |
| TRIM40 | RIG-I, MDA5 | Induces proteasome-mediated degradation | Negative regulation |
| RNF185 | cGAS | Induces IRF3 activation and cytokine production | Positive regulation |
| AMFR | STING | Recruits TBK1 and induces IRF3 activation | Positive regulation |
| USP13 | STING | Inhibits IRF3 activation and cytokine production | Negative regulation |
| USP21 | STING | Inhibits IRF3 activation and cytokine production | Negative regulation |
| USP19 | TAK1 | Inhibits proinflammatory cytokine production | Negative regulation |
Experimental Approaches for Studying K27-Linked Ubiquitination
Methodologies for Chain Synthesis and Characterization
The study of K27-linked ubiquitin chains requires specialized methodologies due to their unique biochemical properties and the absence of dedicated enzymatic synthesis pathways. Non-enzymatic chemical assembly strategies utilizing mutually orthogonal removable amine-protecting groups (Alloc and Boc) enable production of fully natural K27-Ub2 with native isopeptide linkages free of mutations [14]. This approach bypasses the limitations of linkage-specific Ub-conjugating enzymes and ensures structural authenticity.
For structural characterization, solution NMR spectroscopy provides atom-specific information about each ubiquitin unit within the chain [14]. By separately analyzing 15N-enriched distal and proximal ubiquitin units, researchers can quantify amide chemical shift perturbations (CSPs) to identify noncovalent interdomain contacts and conformational changes. Small-angle neutron scattering (SANS) coupled with in silico ensemble modeling further elucidates the dynamic conformational landscapes of K27-linked chains in solution [14] [16].
Functional Assays and Detection Methods
Deubiquitination assays employing multiple DUB families (Cezanne, OTUB1, AMSH, USP2, USP5, Ubp6) provide functional fingerprints for K27-linked chains based on their characteristic resistance to cleavage [14]. This resistance property enables K27-Ub2 to serve as a competitive inhibitor of DUB activity toward other linkages, providing an experimental tool for dissecting DUB specificity and function.
Linkage-specific binding studies using techniques such as pulldown assays with UBA domains and surface plasmon resonance have revealed the unexpected recognition of K27-Ub2 by K48-selective receptors including the UBA2 domain of hHR23a [16]. Mutagenesis studies confirm the structural basis of these interactions, highlighting the versatile recognition capabilities of K27-linked chains.
Diagram 1: Experimental Workflow for K27-Linked Ubiquitin Chain Analysis. This flowchart outlines the integrated approach for synthesizing, characterizing, and functionally validating K27-linked ubiquitin chains, highlighting the multi-technique methodology required to overcome the unique challenges posed by this linkage type.
Advancing research on K27-linked ubiquitination requires specialized reagents and tools designed to address its unique properties and overcome technical challenges.
Table 3: Essential Research Reagents for Studying K27-Linked Ubiquitination
| Reagent/Tool | Function/Application | Key Characteristics |
|---|---|---|
| Chemically Synthesized K29-Ub2 | Reference standard for structural studies | Native isopeptide linkage without mutations; enables biotinylation for detection |
| Linkage-Specific E3 Ligases (TRIM21, TRIM23, RNF185, AMFR) | In vitro and cellular ubiquitination assays | Catalyze formation of K27-linked chains on specific substrates |
| K27-Linkage Resistant DUBs | Negative controls in deubiquitination assays | USP2, USP5, Ubp6 show minimal activity toward K27 linkages |
| NMR with 15N-Labeled Ubiquitin | Structural and dynamic characterization | Reveals chemical shift perturbations and interdomain contacts |
| UBA2 Domain of hHR23a | Binding partner analysis | Recognizes K27-Ub2 through bidentate interactions similar to K48-Ub2 |
| Linkage-Specific Antibodies | Detection and localization in cellular contexts | Enables visualization and quantification of endogenous K27 chains |
| Cryo-EM Sample Preparation | Structural analysis of E3-K27-Ub complexes | Captures transient ubiquitylation transition states with chemical warheads |
K27 Linkages in Context: Comparison with Other Atypical Chains
The functional significance of K27-linked ubiquitination is further illuminated when examined within the broader landscape of atypical ubiquitin chains. While K6-linked chains participate in DNA damage repair and mitophagy, and K11-linked chains regulate the cell cycle and endoplasmic reticulum-associated degradation (ERAD), K27 linkages have carved a specialized niche in immune regulation [15] [19]. K29-linked chains, recently shown to be among the most abundant atypical linkages in eukaryotic cells, function in proteotoxic stress response and cell cycle regulation, particularly enriched in the midbody during mitosis [20]. K33-linked chains mediate protein trafficking and signal transduction of cell surface receptors [15].
What distinguishes K27-linked chains is their particular importance in pathogen defense systems and their unique biochemical properties that include exceptional stability against deubiquitinating enzymes [14]. This specialization highlights the evolutionary adaptation of ubiquitin signaling to meet specific cellular needs, with K27 linkages serving as stable signaling platforms in the context of host-pathogen interactions.
Diagram 2: K27-Linked Ubiquitin Signaling in Antiviral Innate Immunity. This pathway map illustrates how K27-linked ubiquitination regulates multiple nodes in antiviral signaling cascades, from pathogen sensing through transcriptional activation of immune response genes.
K27-linked ubiquitin chains represent a specialized regulatory mechanism within the expanding landscape of atypical ubiquitination, serving as critical modulators of antiviral innate immune signaling and inflammatory pathways. Their unique structural properties, including open conformations and exceptional resistance to deubiquitinating enzymes, enable sustained signaling responses essential for effective pathogen defense. The multifaceted roles of K27 linkages—from regulating nucleic acid sensor pathways to controlling transcriptional activation—highlight their importance as integrative hubs in immune signaling networks.
Future research directions include developing more sensitive and specific tools for detecting endogenous K27-linked chains, elucidating the structural basis for linkage specificity among E3 ligases, and exploring the therapeutic potential of modulating K27-linked ubiquitination in inflammatory diseases and cancer. As part of the broader family of atypical ubiquitin chains, K27 linkages exemplify the sophisticated adaptation of ubiquitin signaling to meet specific cellular needs, offering rich opportunities for scientific discovery and therapeutic innovation.
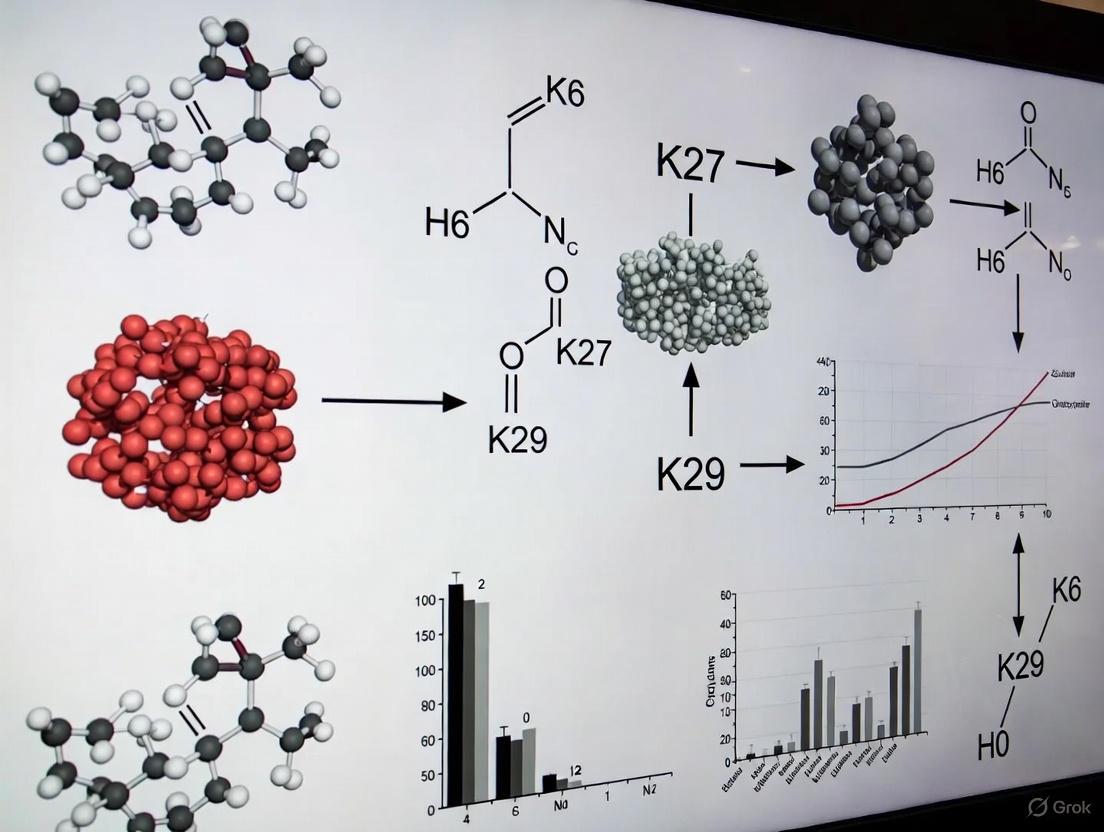
Ubiquitination is a crucial post-translational modification that regulates diverse cellular processes by covalently attaching ubiquitin to target proteins. While K48- and K63-linked ubiquitin chains are well-characterized, the functions of atypical ubiquitin linkages, particularly K29-linked chains, remain less explored. K29-linked ubiquitination represents a non-canonical chain type that has recently emerged as a significant regulator in various cellular pathways beyond protein degradation. This technical review examines the multifaceted roles of K29-linked ubiquitin chains in mRNA stability, kinase regulation, and Wnt/β-catenin signaling, providing researchers with current methodologies and conceptual frameworks for investigating these complex regulatory mechanisms. Understanding K29-linked ubiquitination is essential for deciphering the ubiquitin code and its implications in cellular physiology and disease pathogenesis.
K29-linked ubiquitin chains possess unique structural characteristics that distinguish them from other ubiquitin linkages. Structural analyses reveal that K29-linked diubiquitin adopts an extended conformation with both ubiquitin hydrophobic patches exposed and available for binding interactions [10]. This structural arrangement differs significantly from the compact conformations of K48-linked chains and contrasts with the extended but differently oriented K63-linked chains. The exposed hydrophobic patches enable specific interactions with ubiquitin-binding domains that recognize K29 linkage specificity.
The TRABID zinc finger (NZF) domain exemplifies this specificity, utilizing a binding mode that exploits the inherent flexibility of K29 chains while engaging the hydrophobic patch on only one ubiquitin moiety [10]. This selective recognition mechanism allows K29-linked chains to function as distinct signaling entities in cellular pathways. Furthermore, K29 linkages frequently exist within heterotypic branched chains containing other linkage types, adding complexity to the ubiquitin code and expanding the regulatory potential of ubiquitination [10].
Cellular quantification studies demonstrate that K29-linked chains, while less abundant than K48 or K11 linkages, show tissue-specific enrichment patterns. Notably, contractile tissues such as heart and muscle exhibit relative enrichment of K29 and other atypical chains, suggesting specialized functions in these tissues [21]. This distribution pattern highlights the functional specialization of K29 linkages in specific physiological contexts.
Table 1: Key Characteristics of K29-Linked Ubiquitin Chains
| Characteristic | Description | Functional Implication |
|---|---|---|
| Chain Structure | Extended conformation with exposed hydrophobic patches | Creates unique binding surfaces for specific recognition domains |
| Cellular Abundance | Lower abundance than K48/K11, but tissue-specific enrichment | Specialized rather than universal functions |
| Chain Architecture | Often found in heterotypic/branched chains with other linkages | Increases combinatorial complexity of ubiquitin signaling |
| Recognition Domains | Specific NZF domains (e.g., TRABID) with unique binding modes | Enables specific downstream signaling outcomes |
K29-Linked Ubiquitination in Wnt/β-Catenin Signaling
The Wnt/β-catenin signaling pathway represents a well-characterized system where K29-linked ubiquitination exerts specific regulatory functions. Research has identified that the E3 ligase Smurf1 mediates K29-linked ubiquitination of axin, a critical scaffold component of the β-catenin destruction complex [22]. This modification occurs specifically at lysine residues K789 and K821 of axin and surprisingly does not target axin for proteasomal degradation but instead modulates protein-protein interactions within the Wnt signaling complex.
The functional consequence of Smurf1-mediated K29-linked ubiquitination of axin is the disruption of axin-LRP5/6 interaction, which subsequently attenuates Wnt-stimulated LRP6 phosphorylation and represses Wnt/β-catenin signaling [22]. This non-proteolytic function represents a paradigm shift in understanding how ubiquitination regulates Wnt signaling, moving beyond the traditional degradation-centric view. The identification of specific lysine residues on axin that undergo K29-linked ubiquitination provides mechanistic insight into how this modification interfaces with other post-translational modifications to fine-tune Wnt signaling output.
Table 2: Experimental Evidence for K29-Linked Ubiquitination in Wnt/β-Catenin Signaling
| Experimental Approach | Key Findings | References |
|---|---|---|
| In vitro ubiquitination assay | Smurf1 ubiquitinates axin primarily through K29-linked chains | [22] |
| Site-directed mutagenesis | K789 and K821 identified as critical ubiquitination sites on axin | [22] |
| Co-immunoprecipitation | K29-ubiquitinated axin shows reduced interaction with LRP5/6 | [22] |
| Luciferase reporter assays | Smurf1 overexpression inhibits Wnt/β-catenin signaling | [22] |
| Knockout MEF analysis | Enhanced Wnt signaling in Smurf1-/- murine embryonic fibroblasts | [22] |
Experimental Protocol: Assessing K29-Linked Ubiquitination of Axin
In Vitro Ubiquitination Assay
- Express and purify recombinant human Smurf1 and axin proteins from E. coli using appropriate expression vectors (e.g., pET-28c for Smurf1, pGEX-4T2 for axin)
- Set up ubiquitination reaction in 30 μL mixture containing: 125 ng human E1 enzyme, 500 ng UbcH5c E2 enzyme, 10 μg ubiquitin, 2 μg axin, and 1 μg Smurf1 in reaction buffer (50 mM Tris-HCl pH 7.5, 2 mM ATP, 5 mM MgCl2, 0.5 mM DTT)
- Incubate at 30°C for 2 hours
- Terminate reaction by adding SDS-PAGE loading buffer and analyze by Western blotting
- Detect K29-linked ubiquitination using linkage-specific antibodies (e.g., sAB-K29)
In Vivo Validation
- Co-transfect HEK293T cells with plasmids encoding Smurf1, axin, and wild-type or mutant ubiquitin (K29R)
- Treat cells with MG132 (10 μM, 6 hours) to inhibit proteasomal degradation
- Lyse cells in RIPA buffer supplemented with protease inhibitors and 10 mM N-ethylmaleimide to inhibit deubiquitinases
- Immunoprecipitate axin using specific antibodies
- Analyze ubiquitination by Western blotting with K29-linkage specific antibodies
K29-Linked Ubiquitination in Kinase Regulation and Transcriptional Control
Beyond Wnt signaling, K29-linked ubiquitination plays significant roles in kinase regulation and transcriptional control, particularly during cellular stress responses. Recent research has revealed that the unfolded protein response (UPR) triggers increased K29-linked ubiquitination of the cohesin complex components SMC1A and SMC3 [23]. This specific modification occurs at potentially conserved sites (K1222 on SMC1A) and regulates transcription of cell proliferation-related genes including SERTAD1 and NUDT16L1.
The mechanism involves K29-linked ubiquitination promoting the recruitment of the cohesin release factor WAPL, resulting in cohesin release from chromatin and subsequent transcriptional downregulation [23]. This process effectively inhibits cell proliferation during endoplasmic reticulum stress, allowing cells to conserve resources for stress recovery. The discovery was enabled by CUT&Tag profiling of K29-linked ubiquitin chains in HEK293FT cells, which demonstrated significant overlap with transcriptionally active histone modifications (H3K4me3 and H3K27ac) and accessibility regions identified by ATAC-seq [23].
In kinase regulation, K29-linked ubiquitination exhibits non-proteolytic functions that modulate enzymatic activity and signaling output. The ubiquitin chain-editing complex comprising the HECT E3 ligase UBE3C and the deubiquitinase vOTU has been implicated in the generation and regulation of K29-linked chains that influence kinase activity in various signaling pathways [10].
Figure 1: K29-linked ubiquitination in the unfolded protein response. During ER stress, the UPR triggers K29-linked ubiquitination of cohesin complex components, leading to WAPL-mediated cohesin release and transcriptional repression of proliferation genes.
Methodologies for Studying K29-Linked Ubiquitination
Linkage-Specific Reagents and Detection Methods
Advancements in linkage-specific reagents have been crucial for elucidating K29-linked ubiquitination functions. The development of sAB-K29, a highly specific antibody against K29-linked chains, has enabled precise detection and localization studies [23]. This antibody shows minimal cross-reactivity with other ubiquitin linkage types, making it suitable for various applications including immunofluorescence, Western blotting, and CUT&Tag assays for chromatin-associated ubiquitination.
For structural studies, the K29-selective ubiquitin binding domain from TRABID (NZF1) provides a valuable tool for affinity purification and structural characterization of K29-linked chains [10]. Crystallographic analyses of this domain in complex with K29-diubiquitin have revealed the molecular basis of linkage specificity, informing the design of more specific detection reagents.
Mass spectrometry-based approaches, particularly the Ub-AQUA-PRM (Ubiquitin Absolute Quantification by Parallel Reaction Monitoring) method, enable comprehensive quantification of all ubiquitin chain types in biological samples [21]. This refined assay allows quantification of ubiquitin chain linkages in 10-minute LC-MS/MS runs, facilitating high-throughput screening of ubiquitin chain composition across different tissues and conditions.
Genetic and Cellular Approaches
Genetic studies in model organisms have proven invaluable for understanding K29-linked ubiquitination functions. In S. cerevisiae, genetic interaction screens between gene deletion libraries and lysine-to-arginine ubiquitin mutants have revealed synthetic phenotypes that illuminate the functional relationships between K29 linkages and specific cellular pathways [24].
Cellular studies utilizing DUB-E3 ligase interplay have demonstrated how enzymes like Ubp2, Ubp14, Ufd4, and Hul5 regulate cellular levels of K29-linked unanchored polyubiquitin chains that influence ribosome biogenesis and direct ribosomal proteins to the intranuclear quality control compartment [25]. The accumulation of K29-linked unanchored chains disrupts ribosome assembly and activates the ribosome assembly stress response, connecting K29 ubiquitination to proteostasis maintenance.
Figure 2: Comprehensive workflow for studying K29-linked ubiquitination, encompassing detection methods, functional analysis, and therapeutic exploration.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Essential Research Reagents for Studying K29-Linked Ubiquitination
| Reagent Category | Specific Examples | Applications and Functions |
|---|---|---|
| Linkage-Specific Antibodies | sAB-K29 [23] | Immunofluorescence, Western blotting, CUT&Tag for chromatin-associated K29 chains |
| Ubiquitin Mutants | Ub-K29R [22] [24] | Genetic studies to specifically abrogate K29-linked chain formation |
| E3 Ligase Tools | Smurf1 expression constructs [22], UBE3C/vOTU complex [10] | Enzymatic generation of K29-linked chains for in vitro and in vivo studies |
| Binding Domains | TRABID NZF1 domain [10] | Affinity purification and structural studies of K29-linked chains |
| Mass Spectrometry Standards | AQUA peptides for K29 linkages [21] | Absolute quantification of K29 chain abundance in complex samples |
| Cell Line Models | HEK293FT [23], Smurf1-/- MEFs [22] | Cellular validation of K29-linked ubiquitination functions |
| Deubiquitinase Tools | Ubp2, Ubp14 [25] | Regulation of K29-linked unanchored polyubiquitin chains |
K29-linked ubiquitin chains represent a functionally diverse category of ubiquitin signaling that extends beyond protein degradation to include regulation of Wnt signaling, transcriptional control during stress responses, kinase modulation, and maintenance of proteostasis. The non-proteolytic functions of K29 linkages, particularly in modulating protein-protein interactions and cellular localization, highlight the expanding complexity of the ubiquitin code.
Future research directions should focus on elucidating the full spectrum of E3 ligases and deubiquitinases that specifically regulate K29-linked chains, developing more sophisticated tools for detecting endogenous K29 ubiquitination, and understanding the interplay between K29 linkages and other ubiquitin chain types in heterotypic and branched structures. Furthermore, the therapeutic potential of targeting K29-specific enzymes in disease contexts, particularly cancer and neurodegenerative disorders, warrants increased attention as our understanding of these pathways matures.
The integration of structural biology, quantitative proteomics, and genetic approaches will continue to drive discoveries in this field, potentially revealing novel regulatory mechanisms and therapeutic opportunities for manipulating K29-linked ubiquitination in human disease.
Within the intricate language of cellular signaling, ubiquitination stands out for its extraordinary complexity. While the functions of K48- and K63-linked ubiquitin chains in proteasomal degradation and signal transduction are well-established, the biological roles of atypical ubiquitin linkages (K6, K27, K29) remain a frontier in molecular biology. This whitepaper examines how the distinct structural architectures of these atypical chains directly determine their specialized cellular functions. Through integration of recent biochemical, structural, and proteomic findings, we elucidate the unique properties of K6-, K27-, and K29-linked ubiquitin chains and their implications for cellular regulation and therapeutic development. The emerging paradigm reveals that chain architecture—including linkage type, length, and branching—encodes precise biological information that is decoded by specialized cellular machinery.
Ubiquitin is a 76-amino acid protein that is covalently attached to substrate proteins through a sequential enzymatic cascade involving E1 activating, E2 conjugating, and E3 ligase enzymes [26]. The complexity of ubiquitin signaling arises from the ability of ubiquitin itself to form polymers through its seven lysine residues (K6, K11, K27, K29, K33, K48, K63) or N-terminal methionine (M1) [12]. While K48-linked chains predominantly target substrates for proteasomal degradation and K63-linked chains regulate nonproteolytic processes, the so-called "atypical" chains (K6, K27, K29) exhibit unique structural properties that enable specialized cellular functions [19] [27].
The architectural diversity of ubiquitin chains extends beyond simple linkage type to include chain length and branching patterns [12]. Mixed or branched ubiquitin chains contain more than one type of linkage within the same polymeric structure, dramatically expanding the signaling potential of the ubiquitin code [28]. This review focuses specifically on the structural foundations of K6-, K27-, and K29-linked ubiquitin chains and how their unique architectures dictate biological function in health and disease.
Structural Properties of Atypical Ubiquitin Chains
K27-Linked Ubiquitin Chains
K27-linked ubiquitin chains exhibit exceptional structural and functional properties that distinguish them from all other ubiquitin chain types. Biochemically, K27-linked di-ubiquitin (K27-Ub2) demonstrates remarkable resistance to deubiquitinating enzymes (DUBs), resisting cleavage by linkage-nonspecific DUBs including USP2, USP5, and Ubp6 [14]. This property is unique among all lysine linkages and suggests specialized regulatory functions for K27 chains in contexts requiring stable ubiquitin signals.
Structurally, K27-Ub2 exhibits distinctive characteristics revealed through NMR spectroscopy and small-angle neutron scattering. The proximal ubiquitin unit in K27-Ub2 shows widespread chemical shift perturbations, while the distal ubiquitin exhibits minimal perturbations, indicating an absence of stable noncovalent interdomain contacts [14]. This structural feature likely contributes to the DUB resistance observed in K27 chains and enables unique binding preferences. Surprisingly, despite their structural differences, K27-Ub2 chains are specifically recognized by the UBA2 domain of hHR23a, a receptor typically associated with K48-linked chain recognition [14]. This structural versatility highlights the functional complexity of K27 linkages.
K29-Linked Ubiquitin Chains
K29-linked ubiquitin chains adopt conformations that facilitate specific biological roles in transcriptional regulation and cellular stress response. Chromatin profiling reveals that K29-linked ubiquitin chains are significantly enriched at promoter regions and overlap with transcriptionally active histone modifications including H3K4me3 and H3K27ac [23]. This chromatin association underscores the role of K29 linkages in gene regulation.
During the unfolded protein response (UPR), K29-linked ubiquitination of the cohesin complex increases substantially, with potential modification at the K1222 site on SMC1A [23]. This modification recruits the cohesin release factor WAPL, leading to cohesin release from chromatin and subsequent transcriptional downregulation of cell proliferation-related genes such as SERTAD1 and NUDT16L1 [23]. The structural properties of K29 chains thus enable specific gene regulatory functions during cellular stress.
Chain Length as a Structural Determinant
Beyond linkage type, ubiquitin chain length serves as a critical structural parameter that significantly impacts recognition by ubiquitin-binding proteins. Affinity-based proteomic profiling using length-defined ubiquitin chains has demonstrated that 64-70% of significant interactions with K27-, K29-, and K33-linked chains occur exclusively with long polymers (Ub6+) rather than shorter chains [29]. This pronounced length selectivity adds another layer of specificity to ubiquitin signaling.
Table 1: Structural Properties of Atypical Ubiquitin Chains
| Linkage Type | Structural Features | DUB Sensitivity | Preferred Chain Length | Structural Methods Applied |
|---|---|---|---|---|
| K27 | No stable interdomain contacts; widespread CSPs in proximal Ub | Resistant to most DUBs (USP2, USP5, Ubp6) | Long chains (Ub6+) preferred for most interactions | NMR, SANS, computational modeling |
| K29 | Chromatin-associated; enriched at promoters | Not well characterized | Length-dependent interactions observed | CUT&Tag, ATAC-seq, RNA-seq |
| K6 | Not fully characterized | Variable sensitivity | Not determined | NMR, biochemical assays |
Table 2: Functional Roles of Atypical Ubiquitin Chains
| Linkage Type | Biological Functions | Cellular Processes | Key Enzymes | Specific Substrates |
|---|---|---|---|---|
| K27 | Mitochondrial quality control; proteasome regulation; innate immunity | Mitophagy; protein stabilization | Not well characterized | Miro1 |
| K29 | Transcriptional regulation; ER stress response; cell proliferation control | UPR; gene expression; Wnt signaling | Not well characterized | SMC1A, SMC3 (cohesin complex) |
| K6 | DNA damage repair; mitochondrial regulation | DNA repair pathways; quality control | BRCA1-BARD1 | Unknown substrates |
Experimental Methodologies for Studying Atypical Ubiquitin Chains
Linkage- and Length-Defined Ubiquitin Chain Synthesis
The chemical synthesis of defined ubiquitin chains represents a cornerstone methodology for structural and functional studies. Recent approaches combine click chemistry with gel-eluted liquid fraction entrapment electrophoresis (GELFrEE) to generate ubiquitin chains of defined linkage and length [29]. The methodology involves:
Ubiquitin Monomer Preparation: Generation of bifunctional ubiquitin monomers (Aha75CxPA) containing site-specific modifications at positions x=27, 29, or 33.
Copper-Catalyzed Cycloaddition: Copper(I)-catalyzed alkyne-azide cycloaddition (CuAAC)-mediated protein polymerization with simultaneous desthiobiotin modification.
Chain Length Fractionation: GELFrEE fractionation under denaturing conditions to separate ubiquitin polymers up to the tetramer level in high purity.
Refolding: Dialysis under appropriate conditions to restore native ubiquitin conformation after fractionation.
This methodology enables production of milligram quantities of linkage- and length-defined ubiquitin chains essential for biochemical and structural studies [29].
Chromatin Profiling of Ubiquitin Chains
Cleavage Under Targets and Tagmentation (CUT&Tag) methodology has been adapted to profile the chromatin landscape of atypical ubiquitin chains. The experimental workflow includes:
Cell Permeabilization: Permeabilization of HEK293FT cells to allow antibody access.
Primary Antibody Incubation: Incubation with linkage-specific antibodies (e.g., sAB-K29 for K29-linked chains).
Secondary Antibody Binding: Binding of secondary antibodies conjugated to protein A-Tn5 transposase.
Tagmentation Activation: Magnesium-dependent activation of tagmentation for targeted DNA fragmentation.
Library Preparation and Sequencing: DNA extraction, library preparation, and high-throughput sequencing.
This approach has demonstrated significant enrichment of K29-linked ubiquitin chains at promoter regions, overlapping with transcriptionally active histone marks [23].
Structural Characterization Techniques
Multiple biophysical approaches are employed to characterize the structural properties of atypical ubiquitin chains:
NMR Spectroscopy: Sequential assignment of chemical shifts for both proximal and distal ubiquitin units in di-ubiquitin; quantification of chemical shift perturbations (CSPs) to identify interdomain interactions and conformational changes [14].
Small-Angle Neutron Scattering (SANS): Collection of scattering data under contrast-matched conditions; ensemble modeling to determine conformational distributions and chain compactness [14].
Affinity Enrichment Proteomics: Immobilization of length- and linkage-defined ubiquitin chains on streptavidin agarose; incubation with HEK293T whole cell lysates; LC-MS/MS analysis of enriched proteins; label-free quantification to identify length-selective interaction partners [29].
Functional Implications of Atypical Ubiquitin Chain Architecture
K29-Linked Ubiquitination in Transcriptional Regulation
The architectural properties of K29-linked ubiquitin chains enable specific functions in transcriptional regulation during cellular stress. Under endoplasmic reticulum stress, cells activate the unfolded protein response (UPR), leading to increased K29-linked ubiquitination of the cohesin complex [23]. This modification occurs specifically at promoters of cell proliferation-related genes and recruits the cohesin release factor WAPL, ultimately leading to transcriptional downregulation and inhibition of cell growth [23]. This mechanism demonstrates how K29 chain architecture facilitates gene expression control in response to proteostatic stress.
Branched Ubiquitin Chains in Signal Integration
Branched ubiquitin chains containing atypical linkages represent an emerging dimension of ubiquitin signaling. These complex structures are synthesized through collaborative efforts between E3 ligases with distinct linkage specificities [12]. For example, Ufd4 and Ufd2 collaborate to synthesize branched K29/K48 chains on substrates of the ubiquitin fusion degradation pathway in yeast [12]. Similarly, the APC/C cooperates with UBE2C and UBE2S to form branched K11/K48 chains during mitosis [12].
The functional significance of chain branching lies in its ability to integrate multiple regulatory signals. Branched K48/K63 chains are produced by TRAF6 and HUWE1 during NF-κB signaling, potentially enabling simultaneous engagement of degradative and non-degradative signaling pathways [12]. This architectural complexity allows for sophisticated regulation of protein fate that exceeds the capabilities of homotypic chains.
Research Tools and Reagent Solutions
The study of atypical ubiquitin chains requires specialized reagents and methodologies designed to address their unique structural properties and relative low abundance in cellular contexts.
Table 3: Essential Research Reagents for Studying Atypical Ubiquitin Chains
| Reagent Type | Specific Examples | Applications | Key Features |
|---|---|---|---|
| Linkage-Specific Antibodies | sAB-K29 (K29-specific) | CUT&Tag; immunofluorescence; immunoblotting | High specificity over seven other linkage types |
| Tandem Ubiquitin Binding Entities (TUBEs) | K63-TUBEs; K48-TUBEs; Pan-TUBEs | Affinity enrichment; HTS assays; ubiquitination detection | Nanomolar affinities; protection from DUBs |
| Engineered Ubiquitin Variants | His-tagged Ub; Strep-tagged Ub | Ubiquitinated protein enrichment; proteomic identification | Affinity tag incorporation; mutant ubiquitins |
| Defined Ubiquitin Chains | Triazole-linked K29-Ub2; K27-Ub4 | Structural studies; in vitro assays; interaction profiling | Linkage- and length-defined; DUB-resistant |
| DUB Inhibitors | K27-Ub2 as competitive inhibitor | DUB activity modulation; pathway analysis | Natural DUB resistance of K27 chains |
TUBE-Based Technologies for High-Throughput Applications
Tandem Ubiquitin Binding Entities (TUBEs) have emerged as powerful tools for studying atypical ubiquitination in physiological contexts. These engineered binding proteins consist of multiple ubiquitin-binding domains arranged in tandem, conferring high-affinity interactions with specific ubiquitin chain types [30]. Recent applications include:
High-Throughput PROTAC Screening: K48-specific TUBEs enable rapid assessment of PROTAC-mediated target ubiquitination in 96-well plate formats, facilitating drug discovery efforts [30].
Pathway-Specific Ubiquitination Analysis: K63-specific TUBEs successfully capture inflammatory signaling-induced ubiquitination of RIPK2 following L18-MDP stimulation, demonstrating linkage-specific pathway analysis [30].
Endogenous Protein Ubiquitination Monitoring: TUBE-based platforms overcome limitations of traditional Western blotting by providing quantitative, sensitive detection of endogenous protein ubiquitination without genetic manipulation [30].
The architectural diversity of atypical ubiquitin chains represents a sophisticated mechanism for expanding the functional repertoire of ubiquitin signaling. The unique structural properties of K6-, K27-, and K29-linked chains—including their distinct conformations, chain length preferences, and resistance to deubiquitination—enable precise regulation of cellular processes ranging from transcriptional control to stress adaptation. The emerging understanding of branched ubiquitin chains further demonstrates how linkage mixing creates signaling platforms capable of integrating multiple regulatory inputs.
Future research directions will likely focus on developing more sophisticated tools for probing atypical ubiquitin chain functions in physiological contexts, including improved linkage-specific antibodies, chemical biology approaches for tracing chain dynamics in live cells, and therapeutic strategies targeting disease-relevant ubiquitin signaling nodes. The integration of structural biology with proteomic methodologies will continue to reveal how chain architecture determines biological function in this complex signaling system.
Comparative Abundance and Conservation Across Eukaryotic Species
Ubiquitination is a fundamental post-translational modification that regulates virtually all aspects of eukaryotic cellular physiology. While the roles of canonical ubiquitin linkages (K48 and K63) have been extensively characterized, recent research has unveiled the biological significance of atypical ubiquitin chains, particularly those linked through K6, K27, and K29 residues. These non-canonical linkages exhibit remarkable evolutionary conservation while displaying specialized functions in critical processes ranging from immune signaling to protein quality control. This technical review examines the comparative abundance and conservation patterns of these atypical chains across eukaryotic species, providing researchers with methodological frameworks and conceptual approaches for their continued investigation in both basic science and drug discovery contexts.
Evolutionary Conservation of Ubiquitin and Atypical Linkages
Extreme Sequence Conservation of Ubiquitin
The ubiquitin protein itself displays extraordinary sequence conservation across eukaryotes, with virtual identity observed even between highly divergent species [31]. This extreme conservation stems from both structural and functional constraints:
- Structural stability: Ubiquitin belongs to the beta-grasp fold superfamily, characterized by four or five beta strands forming an anti-parallel sheet and one alpha helix region, creating a compact architecture highly resistant to proteolytic processing and environmental stresses [31].
- Genetic mechanisms: Eukaryotic ubiquitin genes are organized as tandem repeats or fusions with ribosomal proteins, maintained through concerted evolution that prevents drift in redundant copies [31].
- Functional constraints: As a central signaling molecule, ubiquitin must interact with hundreds of different binding partners, limiting tolerable sequence variation [31].
Table 1: Evolutionary Distribution of Ubiquitin System Components
| Component | Evolutionary Origin | Representative Organisms | Genetic Organization |
|---|---|---|---|
| Ubiquitin protein | Last Eukaryotic Common Ancestor | All eukaryotes | Tandem repeats, ribosomal fusions |
| Minimal UPS | Archaea | Caldiarchaeum subterraneum | Operon-like cluster (5 genes) |
| Atypical chain capabilities | Early eukaryotes | Naegleria gruberi, all crown eukaryotes | Distributed genomic loci |
| HECT E3 ligases | Early eukaryotes | Most eukaryotic lineages | Multiple independent genes |
Deep Evolutionary Roots of the Ubiquitin System
Evidence from archaeal and bacterial species reveals that simplified ubiquitin signaling systems predate the emergence of eukaryotes:
- The archaeon Caldiarchaeum subterraneum possesses a minimal ubiquitin system encoded in an operon-like cluster containing single copies of ubiquitin, E1, E2, RING E3, and a deubiquitinase [31].
- The proteome of the amoebo-flagellate Naegleria gruberi, which diverged over one billion years ago, contains more than 100 ubiquitin system genes, indicating early expansion and specialization [31].
- Bacterial homologs of the VTD (Viral Tegument-like Deubiquitinase) family demonstrate that certain deubiquitinase activities emerged prior to eukaryotic radiation [32].
Abundance and Functions of Atypical Ubiquitin Chains
K6-Linked Ubiquitin Chains
K6-linked chains represent a relatively low-abundance ubiquitin linkage with specialized functions:
- DNA damage response: K6-linked chains are generated by the E3 ligase BRCA1 in response to DNA damage and play roles in DNA repair processes [32].
- Mitophagy regulation: Parkin, a mitophagy-associated E3 ligase, generates K6 chains and is itself regulated by K6 ubiquitination during mitochondrial quality control [32].
- Antimicrobial defense: LRSAM1 generates K6 chains during the ubiquitin-dependent autophagy of intracellular bacteria [32].
- Deubiquitination specificity: Certain VTD family deubiquitinases show remarkable specificity for K6-linked chains, suggesting dedicated regulatory pathways [32].
K27-Linked Ubiquitin Chains
K27-linked chains serve as important regulators of innate immune signaling and other processes:
- Innate immune regulation: TRIM23 conjugates K27-linked chains to NEMO, facilitating activation of NFκB and IRF3 transcription factors upon RLR signaling activation [33].
- Signaling platforms: K27 chains on NEMO serve as recruitment platforms for regulatory proteins like Rhbdd3, which recruits the DUB A20 to prevent excessive NFκB activation [33].
- Balanced signaling: K27 linkages appear to balance activation and inhibition in immune pathways, though their complete functional repertoire remains incompletely characterized [33].
K29-Linked Ubiquitin Chains
K29-linked chains have emerged as important players in cellular stress responses and protein degradation:
- Proteotoxic stress: K29-linked chains are associated with cellular responses to proteotoxic stress [34].
- Branched chain formation: TRIP12 generates both K29-linked homotypic chains and K29/K48-branched chains, the latter being associated with targeted protein degradation [34].
- Pathway regulation: K29 linkages participate in diverse cellular pathways including cell division, DNA damage responses, and small-molecule-induced targeted protein degradation [34].
Table 2: Atypical Ubiquitin Chain Functions and Regulatory Enzymes
| Linkage Type | Primary Functions | Forming E3 Ligases | Cleaving DUBs | Relative Abundance |
|---|---|---|---|---|
| K6 | DNA damage response, mitophagy, antimicrobial defense | BRCA1, Parkin, LRSAM1 | VTD family DUBs | Low |
| K27 | Innate immune regulation, signaling platforms | TRIM23 | To be characterized | Low-medium |
| K29 | Proteotoxic stress, targeted degradation | TRIP12, UBR4, UBE3C | To be characterized | Low-medium |
| K11 | Cell cycle regulation, degradation | APC/C, RNF26 | USP19 | Medium |
| K33 | Signaling modulation | To be characterized | To be characterized | Low |
Methodologies for Studying Atypical Ubiquitin Chains
Biochemical and Structural Approaches
Understanding the mechanisms of atypical chain formation requires sophisticated biochemical and structural techniques:
Linkage-Specific Ubiquitin Mutants
- Design principle: Ubiquitin mutants where all lysines except one are mutated to arginine (e.g., ubi-K29) determine whether a specific linkage is sufficient for a process, while single-lysine-to-arginine mutants (e.g., ubi-R29) test whether it is necessary [35].
- Application example: Using ubi-K11 demonstrated that K11-linked chains are sufficient for APC/C-mediated degradation of cyclin B1 and securin, while ubi-R11 impaired this process [35].
Cryo-EM Analysis of E3 Mechanisms
- TRIP12 mechanism: Cryo-EM reveals TRIP12 resembles a pincer, with tandem ubiquitin-binding domains engaging the proximal ubiquitin to position K29 toward the active site, while the HECT domain juxtaposes donor and acceptor ubiquitins [34].
- Geometric constraints: TRIP12 shows exquisite sensitivity to acceptor lysine geometry, with activity abolished when side chains shorter than lysine's tetramethylene linker are tested [34].
Chemical Biology Probes
- Diubiquitin-based photoaffinity probes: Enable profiling of ubiquitin-binding proteins with linkage specificity [36].
- Linkage-specific affimers: Synthetic binding proteins capable of discriminating between different ubiquitin linkage types [36].
- Ubiquitin C-terminal hydrolases: Engineered viral protease-based tools for assessing global ubiquitination architecture [36].
Genetic and Proteomic Methods
Operon Analysis in Prokaryotes
- Identification of operon-like ubiquitin system clusters in archaea like Caldiarchaeum subterraneum provides evolutionary insight into minimal ubiquitin systems [31].
Bioinformatic Discovery of DUB Families
- Iterative profile searches using herpesviral tegument DUB sequences identified the widespread VTD family across eukaryotes and bacteria [32].
- Helitron transposons in zebrafish were found to encode active VTD domains, demonstrating horizontal transfer of DUB capabilities [32].
Branched Chain Analysis
- Collaboration between E3 pairs with distinct specificities (e.g., TRAF6 and HUWE1) produces branched K48/K63 chains during NF-κB signaling [12].
- Sequential ubiquitination first establishes one linkage type before branching E3s recognize this initial mark to add secondary linkages [12].
Visualization of Atypical Ubiquitin Chain Signaling
Collaborative E3 Mechanism for Branched Chain Formation
TRIP12 Mechanism for K29-Linkage Specificity
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Studying Atypical Ubiquitin Chains
| Reagent Category | Specific Examples | Function/Application | Key Features |
|---|---|---|---|
| Linkage-specific ubiquitin mutants | ubi-K29, ubi-R27, ubi-K6-only | Determine necessity and sufficiency of specific linkages | All lysines mutated except one, or single lysine mutations |
| Chemical biology probes | Diubiquitin photoaffinity probes, linkage-specific affimers | Profile ubiquitin-binding proteins and detect specific linkages | Covalent or high-affinity non-covalent linkage recognition |
| Structural biology tools | Cryo-EM transition state mimics, X-ray crystallography | Visualize E3 mechanisms and ubiquitin positioning | Chemical crosslinking to capture transient states |
| Branch-specific reagents | K29/K48-branched diubiquitin probes | Study branched chain recognition and function | Defined branched architecture for biochemical studies |
| Detection reagents | Linkage-specific antibodies, tandem ubiquitin binding entities (TUBEs) | Detect and enrich specific chain types from cell lysates | Selective recognition of unique structural features |
The study of atypical ubiquitin chains has evolved from biochemical curiosities to recognized essential components of eukaryotic cell signaling. The exceptional conservation of K6, K27, and K29 linkages across eukaryotic species underscores their fundamental biological importance, while their relatively low abundance reflects specialized regulatory roles rather than bulk protein modification functions. Future research directions should include:
- Comprehensive profiling of atypical chain abundance across tissue types, subcellular compartments, and physiological states using improved detection methodologies.
- Structural characterization of additional E3 ligases and DUBs specific to atypical linkages to elucidate common and unique mechanistic principles.
- Exploration of crosstalk between different atypical linkages and their combinatorial functions in cellular signaling networks.
- Therapeutic targeting of disease-relevant atypical chain pathways, particularly in neurodegeneration, cancer, and immune disorders where these linkages play important roles.
The continued development of chemical biology tools, structural methods, and genetic approaches will undoubtedly uncover additional functions and regulatory mechanisms for these fascinating ubiquitin signals, potentially opening new avenues for therapeutic intervention in human disease.
Tools of the Trade: Advanced Methodologies for Studying Atypical Ubiquitin Chains
Chemical Biology Approaches for Synthesizing Defined Atypical Ubiquitin Chains
Ubiquitination is a crucial post-translational modification that regulates diverse cellular processes, including protein degradation, DNA repair, and immune signaling. While the functions of K48- and K63-linked ubiquitin chains are well-established, atypical ubiquitin chains (linked via K6, K11, K27, K29, and K33) have emerged as critical regulators in specialized pathways. The study of these atypical linkages has been hampered by significant technical challenges in obtaining homogeneously linked chains of defined length and topology using conventional enzymatic methods. This technical gap has driven the development of innovative chemical biology approaches that provide precise control over ubiquitin chain architecture, enabling researchers to decipher the unique functions of these enigmatic signals in cellular regulation and disease pathogenesis.
Atypical ubiquitin chains play particularly important roles in the regulation of intracellular antiviral innate immune signaling pathways [8]. For instance, K27-linked chains have been implicated in the activation of key immune signaling components: TRIM23-mediated NEMO ubiquitination activates both NFκB and IRF3 pathways, while RNF185-mediated cGAS modification induces IRF3 activation and subsequent type I interferon production [8]. Similarly, K29-linked chains regulate Wnt/β-catenin signaling and mRNA stability, while K33-linked chains influence T-cell receptor signaling and post-Golgi protein trafficking [24]. The functional diversity of these chains underscores the critical need for precise synthetic methods to unravel their distinct biological roles.
Synthesis Methods for Defined Atypical Ubiquitin Chains
Biochemical and Enzymatic Synthesis
Biochemical methods represent the most straightforward approach for synthesizing polyubiquitin chains, utilizing recombinant E1, E2, and/or E3 enzymes to polymerize ubiquitin monomers. These reactions typically yield a statistical mixture of chain lengths, requiring subsequent purification through cation exchange and size exclusion chromatography to isolate defined species [37]. The linkage specificity is determined by careful selection of E2 and E3 enzymes with inherent linkage preferences, though some preparations require additional trimming with linkage-specific deubiquitinases (DUBs) to achieve homogeneity [37].
Table 1: Enzymatic Methods for Atypical Ubiquitin Chain Synthesis
| Linkage Type | E2 Employed | E3 Employed | Post-Synthesis Processing | Key Features |
|---|---|---|---|---|
| K6 | UBE2L3 | NleL | OTUB1 treatment | Associated with DNA damage response and mitophagy |
| K11 | UBE2SΔC | N/A | AMSH treatment | Regulates cell cycle and ERAD; abundant in yeast |
| K27 | Not specified | Not specified | Not established | Resistant to most DUBs; implicated in immune signaling |
| K29 | UBE2L3 | UBE3C | OTUB1, AMSH, and Cezanne | Functions in Wnt signaling and mRNA stability |
| K33 | UBE2D1 | AREL1 | OTUB1 and Cezanne | Regulates T-cell signaling and protein trafficking |
A significant limitation of purely enzymatic approaches is the lack of identified enzymes that specifically generate certain linkage types, particularly K27-linked chains. To overcome this, researchers employ ubiquitin mutants with lysine-to-arginine (K-to-R) substitutions to prevent chain extension at specific positions, combined with C-terminal blocking strategies using modifications like ubiquitin-D77 (Ub-D77), which can be removed by the yeast DUB YUH1 to allow controlled chain elongation [37] [38].
Total Chemical Synthesis
Total chemical synthesis provides exquisite control over ubiquitin chain topology and length, enabling the production of well-defined atypical chains that are difficult to obtain enzymatically. The most common strategy employs native chemical ligation (NCL), which involves chemoselective condensation of peptide thioesters with peptides containing N-terminal cysteine residues [37] [39]. For ubiquitin chain synthesis, this approach utilizes ubiquitin variants containing non-canonical amino acids such as δ-thiolysine or γ-thiolysine at desired linkage sites, which react with C-terminal ubiquitin thioesters to form isopeptide bonds via NCL, followed by chemical desulfurization to yield native isopeptides [37].
The recently developed iso-Ub strategy represents a significant advancement, enabling the synthesis of complex ubiquitin architectures including branched chains. This approach involves synthesizing the portion of the polyubiquitin containing the isopeptides as a single polypeptide chain, defining the linkages and lengths before using NCL as the final step to incorporate additional monomers [37]. This method has successfully delivered defined hexa-ubiquitin chains with branching K11- and K48-linkages, previously a formidable challenge in the field. While chemical synthesis offers unparalleled control, it is time-intensive and results in lower yields (typically <30%) compared to biological approaches, though it provides superior scalability to gram quantities [37].
Semi-Synthetic and Hybrid Approaches
Semi-synthetic methodologies leverage the advantages of both biological and chemical strategies, typically employing recombinant protein expression for most of the structure while utilizing chemical precision for critical steps. The auxiliary-mediated NCL approach uses a photocleavable auxiliary to perform expressed protein ligation (EPL) without requiring a cysteine nucleophile [37]. This auxiliary is removed by photolysis after the S- to N-acyl shift, leaving a native isopeptide linkage. This method has been particularly valuable for producing K27-linked chains, which resist enzymatic synthesis and exhibit unique structural properties and resistance to most deubiquitinases [14].
Genetic code expansion represents another powerful hybrid approach, incorporating non-canonical amino acids through repurposing of the amber stop codon (UAG) in E. coli with an orthogonal tRNA/tRNA synthetase pair. This methodology was used to synthesize K11-K33 branched trimers by incorporating butoxycarbonyl (BOC)-protected lysine at positions K11 and K33 through amber suppression [38]. After Alloc protection of remaining lysines, BOC deprotection, and silver-mediated chemical ligation for branched trimer assembly, the final Alloc deprotection yields the native-like branched ubiquitin chain. This approach has also enabled branched ubiquitin assembly through click chemistry, producing non-hydrolysable chains resistant to DUB activity [38].
Synthesis Method Workflow: Four primary approaches for generating defined atypical ubiquitin chains, each with distinct advantages for specific chain architectures.
Specialized Methods for Branched Ubiquitin Chains
Branched ubiquitin chains represent a particularly complex class of heterotypic ubiquitin signals where at least one ubiquitin moiety is modified at two or more positions simultaneously, creating bifurcation points that significantly expand the ubiquitin code's signaling capacity. The synthesis of defined branched chains requires specialized approaches that combine multiple enzymatic and chemical strategies.
The most common method for generating branched ubiquitin trimers utilizes C-terminally blocked proximal ubiquitin (Ub1-72 or UbD77), with mutant distal ubiquitins ligated sequentially using specific enzymes for each desired linkage [38]. For example, branched K48-K63 trimers can be assembled by first generating a K63 dimer from Ub1-72 and UbK48R,K63R using UBE2N and UBE2V1, followed by K48 linkage of UbK48R,K63R to the proximal Ub1-72 using K48-specific enzymes like UBE2R1 or UBE2K [38]. To enable assembly of more complex tetrameric branched structures, the Ub-capping approach uses the yeast DUB Yuh1 to trim the C-terminus of a D77-blocked ubiquitin, or the M1-specific DUB OTULIN to remove proximal caps after branch formation, thereby exposing the native C-terminus for further chain extension [38].
Recently, a photo-controlled enzymatic assembly method was developed using chemically synthesized ubiquitin moieties where target lysine residues are protected by photolabile 6-nitroveratryloxycarbonyl (NVOC) groups [38]. Through alternating cycles of linkage-specific elongation, NVOC deprotection with UV irradiation, and subsequent elongation, this approach enables assembly of complex branched architectures like K48-K63 branched tetramers using wildtype ubiquitin, representing a significant advancement in the field.
Table 2: Experimentally Confirmed Branched Ubiquitin Chain Architectures
| Branch Linkages | Synthetic Enzymes/Methods | Biological Functions | Structural Features |
|---|---|---|---|
| K11/K48 | APC/C (UBE2C + UBE2S) | Cell cycle regulation, proteasomal degradation | Closed conformation promoting proteasomal recognition |
| K29/K48 | Ufd4 + Ufd2 collaboration | Ubiquitin fusion degradation pathway | Specific recognition by UBA-UBL domain proteins |
| K48/K63 | TRAF6 + HUWE1 collaboration | NF-κB signaling, apoptotic regulation | Conversion from non-degradative to degradative signal |
| K6/K48 | Parkin, NleL | Mitophagy, mitochondrial quality control | Parkin mutations associated with Parkinson's disease |
| K11/K33 | Genetic code expansion + chemical ligation | Experimental model for branched chain recognition | Synthetic architecture demonstrating methodology |
Experimental Protocols for Key Applications
Protocol: Enzymatic Synthesis of K11-Linked Diubiquitin
This protocol describes the synthesis of K11-linked diubiquitin using the E2 enzyme UBE2SΔC, based on established methodologies with modifications for atypical chain production [37].
Recombinant Protein Expression: Express and purify ubiquitin (wild-type and K11-only mutant Ub(K0) with all lysines except K11 mutated to arginine) and the E2 enzyme UBE2SΔC (residues 1-200) from E. coli using standard nickel-affinity and ion-exchange chromatography.
Enzymatic Reaction: Prepare a 5 mL reaction containing 50 mM Tris-HCl (pH 7.5), 10 mM MgCl₂, 0.5 mM DTT, 5 mM ATP, 10 μM human E1 enzyme, 100 μM UBE2SΔC, and 1 mM Ub(K0) mutant. Incubate at 30°C for 4 hours.
Reaction Monitoring: Analyze reaction progress by SDS-PAGE and anti-ubiquitin immunoblotting. The reaction is complete when most Ub(K0) migrates at the molecular weight corresponding to diubiquitin.
Purification: Terminate the reaction by adding 1% trifluoroacetic acid (TFA). Purify K11-linked diubiquitin by cation exchange chromatography (Resource S column) using a 0-500 mM NaCl gradient in 50 mM sodium acetate (pH 4.5). Further purify by size exclusion chromatography (Superdex 75) in 20 mM Tris-HCl (pH 7.5), 150 mM NaCl.
Validation: Verify chain linkage by mass spectrometry and immunoblotting with linkage-specific antibodies. Treat with the K11-linkage specific DUB Cezanne as an additional validation step [37].
Protocol: Semi-Synthesis of K27-Linked Diubiquitin Using Auxiliary-Mediated Ligation
This protocol describes the production of K27-linked diubiquitin through a semi-synthetic approach that addresses the lack of specific E3 ligases for this linkage type [37] [14].
Ubiquitin Activation: Prepare ubiquitin thioester by intein-mediated expression in E. coli using a ubiquitin-intein-chitin binding domain fusion. Cleave with mercaptoethanesulfonate (MESNA) to generate ubiquitin-MESNA thioester.
Auxiliary Preparation: Chemically synthesize a ubiquitin fragment (residues 1-73) containing a C-terminal thioester and a ubiquitin fragment (residues 27-76) containing an N-terminal cysteine and a photocleavable auxiliary at K27.
Ligation Reaction: Combine ubiquitin-MESNA thioester (1.2 equiv) with the ubiquitin K27-auxiliary fragment (1.0 equiv) in ligation buffer (6 M guanidine hydrochloride, 100 mM sodium phosphate, 30 mM MPA, pH 7.2). Incubate at 37°C for 12-16 hours.
Photocleavage: Dilute the ligation product and irradiate at 365 nm for 2 hours to remove the photocleavable auxiliary, revealing the native isopeptide linkage.
Refolding and Purification: Dialyze against refolding buffer (20 mM Tris-HCl, pH 7.5) and purify by ion-exchange and size-exclusion chromatography. Verify the product by mass spectrometry and NMR spectroscopy [14].
K27 Diubiquitin Synthesis: Specialized semi-synthetic approach required for K27-linked chains due to their resistance to enzymatic production and unique structural properties.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Essential Research Reagents for Atypical Ubiquitin Chain Synthesis and Analysis
| Reagent/ Tool | Function/Application | Key Features and Examples |
|---|---|---|
| Linkage-Specific E2 Enzymes | Catalyze formation of specific ubiquitin chain linkages | UBE2S (K11-specific), UBE2L3 (K6 and K29), UBE2D1 (K33) [37] |
| DUBs for Linkage Validation | Confirm linkage specificity through selective cleavage | Cezanne (K11-specific), OTUB1 (K48-specific), AMSH (K63-specific) [14] |
| Ubiquitin Mutants | Control chain elongation and branching patterns | Ub(K0) variants with single lysine; K-to-R mutations to block specific linkages [37] [38] |
| Chemical Biology Probes | Enable specific conjugation and detection | Thiol-ene coupling reagents, photo-cleavable auxiliaries, click chemistry components [37] [38] |
| Linkage-Specific Antibodies | Detect specific ubiquitin linkages in assays | Commercial antibodies for K11, K27, K29, K33 linkages (validation required) |
| NMR Isotope Labels | Study chain structure and dynamics | 15N-, 13C-labeled ubiquitin for solution NMR spectroscopy [14] |
Chemical biology approaches have revolutionized our ability to synthesize defined atypical ubiquitin chains, providing essential tools to decipher their unique structural and functional properties. The integration of enzymatic methods with sophisticated chemical strategies has enabled researchers to overcome the limitations of natural enzymatic machinery, particularly for challenging targets like K27-linked and branched ubiquitin chains. These well-defined reagents have revealed the critical importance of atypical chains in specialized cellular processes, including immune signaling, mitochondrial quality control, and transcriptional regulation. As these synthetic methodologies continue to advance, they will undoubtedly uncover new dimensions of the ubiquitin code, potentially revealing novel therapeutic targets for diseases ranging from neurodegeneration to cancer. The ongoing development of more efficient and versatile synthesis platforms promises to further accelerate our understanding of these complex post-translational modifications and their multifaceted roles in cellular regulation.
Protein ubiquitination, a pivotal post-translational modification, regulates virtually all essential eukaryotic cellular processes. The complexity of ubiquitin (Ub) signaling extends far beyond simple monoubiquitination, encompassing diverse polyubiquitin chains where ubiquitin molecules are linked through one of seven lysine residues (K6, K11, K27, K29, K33, K48, K63) or its N-terminal methionine (M1). These linkages form distinct chain topologies that are interpreted as a "ubiquitin code," dictating specific cellular outcomes for modified substrates, from proteasomal degradation to activation of immune signaling pathways [1] [36]. While K48- and K63-linked chains are the most well-characterized, the so-called "atypical" linkages—including K6, K27, and K29—have emerged as critical regulators in processes such as mitophagy, innate immune response, and proteotoxic stress, though their study has been hampered by technical challenges [33] [14] [34].
A primary obstacle in ubiquitin research, particularly for atypical linkages, is their low abundance relative to canonical chains and the lability of ubiquitin modifications due to active deubiquitinases (DUBs) in cell lysates [40]. Affinity-based enrichment techniques have therefore become indispensable tools, enabling researchers to overcome these barriers by selectively isolating ubiquitinated proteins or specific chain types from complex biological samples prior to downstream analysis by immunoblotting or mass spectrometry. This technical guide provides an in-depth examination of the three cornerstone affinity-based separation platforms—antibodies, ubiquitin-binding domains (UBDs), and TUBEs (tandem-repeated ubiquitin-binding entities)—with a specific focus on their application in elucidating the functions of K6, K27, and K29 ubiquitin linkages.
Core Technologies for Ubiquitin Enrichment
Ubiquitin-Binding Domains (UBDs) and TUBEs
Ubiquitin-binding domains are modular protein domains that recognize non-covalent surfaces on ubiquitin. Their inherent specificity for ubiquitin makes them powerful tools for affinity purification. The following table summarizes key UBD-based tools used in ubiquitin enrichment.
Table 1: Key UBD-Based Tools for Ubiquitin Enrichment
| Tool Name | Class/Type | Key Features | Applications in Atypical Chain Research |
|---|---|---|---|
| OtUBD [41] | Single, high-affinity UBD from O. tsutsugamushi | - Strong enrichment of mono- and poly-Ub proteins- Compatible with native & denaturing workflows- Versatile resin compositions and elution methods | - Protocol used to enrich ubiquitinated proteins from yeast and mammalian lysates for ubiquitinome analysis. |
| Tandem Hybrid UBD (ThUBD) [40] | Artificial tandem UBD construct | - Enhanced affinity and avidity- Recognizes eight types of Ub chains with high efficiency and reduced bias- Couples with DRUSP method for superior performance | - Deep ubiquitinome profiling of mouse liver fibrosis models.- Extracts ~3x stronger ubiquitin signal versus control methods. |
| TUBEs [36] | Tandem-repeated Ub-Binding Entities | - Multiple UBDs in series confer high avidity- Protects ubiquitinated proteins from DUBs and proteasomal degradation- Broad specificity for diverse ubiquitin linkages | - Global analysis of ubiquitination; tracking and quantifying endogenous Ub architectures.- Often coupled with proteomic analysis. |
Linkage-Specific Antibodies
Antibodies provide the highest degree of specificity for detecting particular ubiquitin linkages. Recent advances have yielded reagents capable of distinguishing the unique structural topologies of atypical chains.
Table 2: Linkage-Specific Antibodies and Reagents
| Specificity | Reagent Type | Key Characteristics and Findings |
|---|---|---|
| K27 Linkage | Affimers, Synthetic Antigen Binders [36] | - K27 linkages exhibit unique resistance to most deubiquitinases (DUBs).- Structural studies show K27-linked di-ubiquitin (K27-Ub2) may be specifically recognized by proteasomal shuttle protein hHR23a. |
| K29 Linkage | Linkage-Selective Reagents [36] | - K29-linked chains are associated with proteotoxic stress responses.- TRIP12 E3 ligase specifically forges K29 linkages and K29/K48-branched chains. |
| N-Terminal Ubiquitination | Anti-GGX Monoclonal Antibodies [42] | - Four unique mAbs (1C7, 2B12, 2E9, 2H2) selectively recognize N-terminal diglycine (GG) remnants on tryptic peptides.- Crucially, they do not cross-react with isopeptide-linked GG on lysine (K-ε-GG).- Solved structure of 1C7 Fab bound to a Gly-Gly-Met peptide reveals molecular basis for selectivity. |
The following diagram illustrates the core workflow and logical relationship between different affinity-based enrichment methods and their downstream applications, highlighting the critical decision points for researchers.
Detailed Experimental Protocols
OtUBD-Based Enrichment of Ubiquitinated Proteins
The OtUBD protocol provides a versatile method for enriching ubiquitinated proteins from crude lysates, adaptable for both native and denaturing conditions to answer different biological questions [41].
Materials & Reagents:
- OtUBD Affinity Resin: Prepared by coupling the recombinant OtUBD protein to a chosen chromatography resin (e.g., CNBr-activated Sepharose).
- Lysis Buffers:
- Native Lysis Buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 1 mM DTT, supplemented with protease inhibitors and DUB inhibitors (e.g., 10 mM N-ethylmaleimide).
- Denaturing Lysis Buffer: 50 mM Tris-HCl (pH 7.5), 1% SDS, 1 mM DTT.
- Wash Buffers:
- High-Salt Wash: 50 mM Tris-HCl (pH 7.5), 500 mM NaCl, 0.5% NP-40.
- Low-Salt Wash: 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 0.5% NP-40.
- Elution Buffer: 1X SDS-PAGE sample buffer, or 100 mM glycine (pH 2.5) for neutralization and dialysis.
Step-by-Step Procedure:
- Lysate Preparation:
- For native workflow (preserves non-covalent interactions): Prepare lysates using Native Lysis Buffer. Clarify by centrifugation at 15,000 x g for 15 minutes at 4°C.
- For denaturing workflow (analyzes covalent ubiquitinome only): Lyse cells in Denaturing Lysis Buffer. Heat lysates at 95°C for 5-10 minutes to denature proteins. Dilute 10-fold with a neutral buffer without SDS before proceeding to reduce SDS concentration.
- Pulldown: Incubate the clarified lysate with OtUBD affinity resin for 2-4 hours at 4°C with gentle end-over-end mixing.
- Washing: Pellet the resin and wash sequentially.
- Wash twice with High-Salt Wash Buffer.
- Wash twice with Low-Salt Wash Buffer.
- A final wash with a salt-free buffer (e.g., 50 mM Tris-HCl, pH 7.5) can be performed.
- Elution: Elute bound ubiquitinated proteins by boiling the resin in 1X SDS-PAGE sample buffer for 5-10 minutes, or by using a low-pH elution buffer.
- Downstream Analysis: The eluate can now be analyzed by immunoblotting with linkage-specific antibodies or prepared for liquid chromatography-tandem mass spectrometry (LC-MS/MS) for ubiquitinome profiling.
DRUSP (Denatured-Refolded Ubiquitinated Sample Preparation) with ThUBD
The DRUSP method is an innovative workflow designed to overcome major limitations of native lysate preparations, such as incomplete protein extraction and DUB activity, thereby significantly enhancing the sensitivity and reproducibility of ubiquitinomics [40].
Materials & Reagents:
- Strong Denaturation Buffer: 8 M Urea, 50 mM Tris-HCl (pH 8.0), 1% SDS.
- Refolding Buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5% NP-40.
- Tandem Hybrid UBD (ThUBD) Resin: Affinity resin coupled with the engineered ThUBD.
- Centrifugal Filter Units: with appropriate molecular weight cut-off for buffer exchange.
Step-by-Step Procedure:
- Denatured Extraction: Lyse cells or tissue directly in a Strong Denaturation Buffer. This effectively inactivates DUBs and proteasomes and maximizes the extraction of insoluble ubiquitinated proteins.
- Refolding via Buffer Exchange: Dilute the denatured lysate and transfer it to a centrifugal filter unit. Perform sequential centrifugation and dilution with Refolding Buffer to remove denaturants (urea, SDS) and allow the ubiquitinated proteins to refold into a native conformation. This step is crucial for restoring the native spatial structure of ubiquitin and ubiquitin chains, making them recognizable by the ThUBD.
- ThUBD Enrichment: Incubate the refolded lysate with the ThUBD resin. The ThUBD exhibits high affinity for eight types of ubiquitin chains with minimal bias.
- Washing and Elution: Wash the resin thoroughly to remove non-specifically bound proteins. Elute the enriched ubiquitinated proteins for downstream LC-MS/MS analysis.
- MS Proteomics: The eluate is digested with trypsin. The resulting peptides, including those with the characteristic diglycine (GG) remnant on lysine, are analyzed by LC-MS/MS to map ubiquitination sites.
Enrichment of N-Terminally Ubiquitinated Substrates
This protocol uses specific anti-GGX monoclonal antibodies to isolate and identify proteins modified by the non-canonical N-terminal ubiquitination [42].
Materials & Reagents:
- Anti-GGX mAbs: Clones 1C7, 2B12, 2E9, or 2H2.
- Cell Line: Optional inducible UBE2W (the E2 for N-terminal ubiquitination) overexpression system.
- Lysis & IP Buffer: Standard RIPA or similar IP-compatible buffer.
- Trypsin: Mass spectrometry grade.
Step-by-Step Procedure:
- Generate Peptide Library: Prepare a tryptic digest from cell lysates. This cleaves proteins after lysine and arginine residues, generating peptides. For an N-terminally ubiquitinated protein, trypsin cleavage produces a peptide with a diglycine motif at its N-terminus (GGX-peptide).
- Immunoaffinity Enrichment: Incubate the tryptic peptide mixture with anti-GGX mAbs (e.g., 1C7) conjugated to beads. These antibodies are highly selective for the linear N-terminal GG motif and show no cross-reactivity with the isopeptide-linked K-ε-GG remnant from canonical ubiquitination.
- Washing: Wash the beads extensively to remove non-specifically bound peptides.
- Elution and LC-MS/MS: Elute the enriched GGX-peptides and analyze them by LC-MS/MS. This allows for the identification of the protein and the exact site of N-terminal ubiquitination.
- Validation: Candidate substrates, such as UCHL1 and UCHL5 identified in the original study, can be validated biochemically to confirm the functional consequences of N-terminal ubiquitination.
Application in Atypical Ubiquitin Chain Research
Elucidating Signaling Pathways Regulated by Atypical Chains
Affinity-based tools have been instrumental in uncovering the roles of atypical chains in critical signaling pathways, particularly in the regulation of antiviral innate immunity and proteostasis. The following diagram summarizes key pathways regulated by K27 and K29 linkages.
K27-Linked Chains in Immune Signaling: The use of linkage-specific tools has revealed that K27-linked chains are important regulators of the antiviral innate immune response. For example:
- TRIM23 conjugates K27-linked chains to NEMO (a component of the IKK complex), which is required for the activation of IRF3 and NF-κB transcription factors upon viral detection [33].
- The linear ubiquitin chain assembly complex (LUBAC), known for forming M1-linked chains, also contributes to immune regulation. Hepatitis B virus can recruit LUBAC to the mitochondrial antiviral-signaling protein (MAVS), leading to the formation of linear chains that disrupt the MAVS signalosome and inhibit the type I interferon response [33].
K29-Linked Chains in Degradation and Stress:
- RNF26 utilizes K11-linked chains to regulate the stability of STING, but K29-linked chains are implicated in other degradation-related pathways, such as the ubiquitin fusion degradation (UFD) pathway [33] [12].
- TRIP12 is a HECT E3 ligase that specifically generates K29-linked homotypic chains and K29/K48-branched chains. These chains are associated with the cellular response to proteotoxic stress and are linked to neurodegenerative disorders and autism spectrum disorders [34]. The unique geometry of the TRIP12 active site, which precisely positions the acceptor ubiquitin's K29 residue, ensures linkage specificity.
Research Reagent Solutions Toolkit
The following table compiles essential reagents for designing experiments focused on affinity-based separation of ubiquitinated proteins.
Table 3: Essential Research Reagent Solutions for Ubiquitin Enrichment
| Category | Reagent | Specific Function/Application |
|---|---|---|
| Affinity Resins | OtUBD Affinity Resin [41] | Core matrix for versatile (native/denaturing) enrichment of mono- and poly-Ub proteins. |
| Tandem Hybrid UBD (ThUBD) Resin [40] | High-performance resin for deep ubiquitinome profiling with minimal linkage bias. | |
| Specialized Antibodies | Anti-K27 Linkage Reagents [36] | Affimers or synthetic binders for specific detection/enrichment of K27-linked chains. |
| Anti-GGX mAbs (1C7, 2B12, etc.) [42] | Selective enrichment of N-terminally ubiquitinated peptides from tryptic digests for MS. | |
| Buffers & Kits | DRUSP-Compatible Buffers [40] | Denaturing and refolding buffer systems for optimal protein extraction and UBD recognition. |
| DUB Inhibitor Cocktails | Essential additive to native lysis buffers to preserve ubiquitin signals during preparation. | |
| Critical Cell Lines/Tools | UBE2W-Inducible System [42] | Tool for studying N-terminal ubiquitination by overexpressing the key E2 enzyme. |
| TRIP12 Recombinant Enzyme [34] | Key E3 ligase for in vitro studies of K29-linked and K29/K48-branched chain synthesis. |
Affinity-based separation technologies form the backbone of modern ubiquitin research. The continued refinement of UBDs like OtUBD and ThUBD, the development of highly specific antibodies for atypical linkages and non-canonical modifications, and the implementation of robust protocols like DRUSP have collectively empowered scientists to dissect the complex functions of the ubiquitin code with unprecedented precision. As these tools evolve, they will undoubtedly unlock further secrets of K6, K27, K29, and other atypical ubiquitin chains, illuminating their roles in health and disease and opening new avenues for therapeutic intervention.
Mass Spectrometry-Based Strategies for Ubiquitinomics and Linkage Mapping
Protein ubiquitination, a fundamental post-translational modification, regulates nearly every cellular process in eukaryotes, from protein degradation to immune signaling and DNA repair. While the roles of canonical K48- and K63-linked ubiquitin chains are well-established, recent research has unveiled the critical functions of atypical ubiquitin linkages—specifically K6, K27, and K29—in specialized biological pathways. These atypical chains represent a sophisticated regulatory layer in cellular signaling, with distinct structural properties and cellular functions that are only beginning to be understood. The study of these chains has been hampered by technical challenges, including their low abundance in cells and the difficulty in detecting and characterizing them amid a complex background of abundant canonical chains.
Mass spectrometry-based proteomics has emerged as an indispensable tool for deciphering the ubiquitin code, enabling researchers to identify ubiquitinated proteins, map modification sites, and determine polyubiquitin chain linkages. This technical guide explores cutting-edge mass spectrometry strategies specifically designed for comprehensive ubiquitinomics and linkage mapping, with particular emphasis on their application to the functionally enigmatic K6, K27, and K29 chain types. We focus on the most recent methodological advances that are pushing the boundaries of what can be detected and quantified in the atypical ubiquitin landscape.
Methodological Landscape in Ubiquitinomics
The evolution of mass spectrometry technologies has progressively enhanced our ability to study the ubiquitinome. Early approaches relied heavily on data-dependent acquisition (DDA) methods and shotgun proteomics, which provided the first large-scale views of ubiquitinated proteins. These methods enabled the identification of thousands of ubiquitination sites but faced limitations in reproducibility, quantification accuracy, and coverage depth, particularly for low-abundance atypical chain signatures.
The field has since witnessed a paradigm shift toward data-independent acquisition (DIA) methods, which offer significantly improved reproducibility and quantification precision. When coupled with advanced computational approaches like deep neural network-based processing (exemplified by DIA-NN software), DIA methods have dramatically expanded ubiquitinome coverage, enabling identification of up to 70,000 ubiquitinated peptides in single MS runs compared to approximately 21,000 with DDA—representing a more than threefold increase in coverage [43].
Table 1: Comparison of Key Ubiquitinomics Mass Spectrometry Approaches
| Method | Typical Identifications | Quantitative Precision | Key Advantages | Best Applications |
|---|---|---|---|---|
| Data-Dependent Acquisition (DDA) | ~21,000 K-GG peptides | Moderate (CV ~15-20%) | Established workflows, extensive literature | Targeted studies, verification experiments |
| Data-Independent Acquisition (DIA) | ~68,000 K-GG peptides | High (CV ~10%) | Excellent reproducibility, minimal missing values | Large-scale temporal studies, comprehensive profiling |
| Top-Down MS | Intact protein analysis | N/A | Preserves linkage information and proteoforms | Direct mapping of ubiquitin chain architecture |
| DIA with Neural Network Processing | ~70,000 K-GG peptides | Very High (CV <10%) | Maximized coverage and quantification accuracy | Discovery studies, mode-of-action investigations |
For the specific challenge of mapping ubiquitin chain linkages, including the atypical K6, K27, and K29 connections, two complementary approaches have emerged: bottom-up methods that infer linkage types from signature peptides or di-glycine remnants, and top-down strategies that analyze intact ubiquitinated proteins or polyubiquitin chains. Top-down mass spectrometry (TD-MS) represents a particularly promising frontier, as it preserves the intact ubiquitin chain architecture, allowing simultaneous determination of ubiquitination sites and chain topology on protein substrates [44].
Analytical Workflows for Ubiquitinomics
Sample Preparation and Lysis Optimization
The foundation of successful ubiquitinomics begins with appropriate sample preparation. Recent methodological comparisons have demonstrated that sodium deoxycholate (SDC)-based lysis buffers, supplemented with chloroacetamide (CAA) for immediate cysteine protease inhibition, outperform conventional urea-based buffers. This SDC-based approach increases ubiquitin site coverage by approximately 38% while maintaining high enrichment specificity [43]. The protocol involves immediate boiling of samples after lysis in SDC buffer containing high concentrations of CAA, which rapidly inactivates deubiquitinating enzymes (DUBs) that would otherwise erase ubiquitination signals during sample processing. This step is particularly critical for preserving the labile signals of atypical ubiquitin chains, which may be turned over more rapidly than their canonical counterparts.
Following lysis, proteins are digested with trypsin, which cleaves after lysine and arginine residues. This digestion generates a characteristic di-glycine (K-GG) remnant—a 114.0429 Da mass tag left on the modified lysine ε-amino group—that serves as a signature for ubiquitination sites. K-GG-containing peptides are then enriched using immunoaffinity purification with specific antibodies before mass spectrometry analysis. For linkage-specific analyses, alternative proteases such as Lys-C may be employed to generate longer ubiquitin-derived peptides that retain information about the linkage type between ubiquitin monomers.
Mass Spectrometry Acquisition and Data Processing
For comprehensive ubiquitinome profiling, the current state-of-the-art employs data-independent acquisition (DIA) with optimized liquid chromatography gradients and mass spectrometry parameters. A typical workflow uses medium-length (75-125 minute) nanoLC gradients coupled to high-resolution mass spectrometers. The DIA approach systematically fragments all ions within predefined m/z windows across the entire chromatographic elution profile, ensuring comprehensive and reproducible detection of ubiquitin remnant peptides [43].
Data processing represents a critical step in the workflow. The DIA-NN software package, with its deep neural network-based processing and specialized scoring module for modified peptides, has demonstrated particular efficacy for ubiquitinomics applications. When operated in "library-free" mode (searching directly against sequence databases without requiring experimental spectral libraries), DIA-NN can identify and quantify approximately 68,000 K-GG peptides with a median coefficient of variation below 10% across replicates [43]. This represents a significant advancement over traditional DDA approaches, which typically yield higher rates of missing values in replicate analyses.
Figure 1: Comprehensive DIA-MS Workflow for Ubiquitinomics
Specialized Approaches for Linkage Mapping
Top-Down Strategies for Direct Linkage Determination
While bottom-up ubiquitinomics excels at comprehensive site identification, it typically loses information about the connectivity between ubiquitin monomers in polyubiquitin chains. To address this limitation, top-down mass spectrometry approaches have been developed that analyze intact ubiquitinated proteins or polyubiquitin chains. The recently developed UbqTop computational platform utilizes a Bayesian-like scoring algorithm to predict ubiquitin chain topology directly from tandem MS (MS2) fragmentation data of intact ubiquitinated species [44].
This integrated strategy enables simultaneous determination of the ubiquitin modification site and the architecture of the attached ubiquitin chain. To manage the analytical challenge posed by complex protein substrates, researchers often combine this with selective Asp-N proteolysis, which digests the protein substrate while preserving intact ubiquitin chains. This hybrid proteolysis-TD-MS approach enables direct, site-resolved mapping of ubiquitin chain topology on proteins and has demonstrated utility for resolving isomeric chains and branched architectures involving atypical linkages [44].
Advanced Methods for Branched Chain Analysis
The ubiquitin code increases in complexity with the formation of branched ubiquitin chains, in which one or more ubiquitin monomers are simultaneously modified on at least two different acceptor sites. These branched architectures include combinations such as K11/K48, K29/K48, and K48/K63, with evidence for atypical linkage combinations including K6/K11, K6/K48, K27/K29, and K29/K33 [12].
Branched chains are often synthesized through the collaboration of E3 ligase pairs with distinct linkage specificities. For example, during the ubiquitin fusion degradation (UFD) pathway in yeast, Ufd4 and Ufd2 collaborate to synthesize branched K29/K48 chains on substrates [12]. Similarly, in NF-κB signaling, TRAF6 and HUWE1 cooperate to produce branched K48/K63 chains [12]. Mapping these complex structures requires specialized analytical approaches, including the use of linkage-specific antibodies, ubiquitin-binding domains (UBDs), and advanced fragmentation techniques that can preserve and reveal the connectivity information within these branched polymers.
Table 2: Essential Research Reagents for Ubiquitinomics and Linkage Mapping
| Reagent/Category | Specific Examples | Function/Application | Considerations for Atypical Linkages |
|---|---|---|---|
| Lysis Buffers | SDC buffer + CAA | Protein extraction with DUB inhibition | Superior to urea for preserving K27 chains |
| Proteases | Trypsin, Asp-N | Generate signature peptides | Asp-N preserves Ub chains for top-down MS |
| Enrichment Reagents | K-GG antibodies | Immunoaffinity purification of ubiquitinated peptides | Efficiency varies for different linkage types |
| DUB Inhibitors | MG-132 (proteasome) | Stabilize ubiquitinated proteins | Affects degradative vs non-degradative signals |
| Computational Tools | UbqTop, DIA-NN | Data processing and linkage prediction | Specialized algorithms for atypical chains |
| Linkage Standards | Recombinant di-Ub chains | Method calibration and validation | Essential for K6, K27, K29 reference spectra |
Functional Roles of Atypical Ubiquitin Linkages
Biological Significance of K6, K27, and K29 Chains
The atypical ubiquitin linkages K6, K27, and K29 play specialized roles in cellular regulation, distinct from the well-characterized degradative (K48) and signaling (K63) functions of canonical chains. K27-linked ubiquitination has emerged as a key regulator of innate immune signaling, with multiple E3 ligases including TRIM23, TRIM26, and RNF185 conjugating K27-linked chains to substrates like NEMO, MAVS, and cGAS to modulate antiviral responses and inflammatory pathways [8]. Structurally, K27-linked di-ubiquitin (K27-Ub2) exhibits unique characteristics, including unusual resistance to deubiquitinating enzyme (DUB) cleavage and open conformational states in solution that facilitate bidentate interactions with ubiquitin receptors like the UBA2 domain of hHR23A [16].
K29-linked chains have been implicated in diverse processes including mRNA stability regulation through modification of the RNA-binding protein HuR, and in the regulation of the anaphase-promoting complex (APC) [24]. Meanwhile, K6-linked ubiquitination functions in DNA damage response through the BRCA1-BARD1 E3 ligase complex and in mitophagy through Parkin-mediated signaling [24]. These chains are often embedded in more complex branched architectures, such as the K27/K29 branched chains synthesized by RNF34 on MAVS to induce autophagy-mediated degradation and restrict type I interferon response during antiviral immunity [8].
Spatial Regulation of Ubiquitin Signaling
Recent advances in subcellular proteomics have revealed that ubiquitin signaling is compartmentalized within cells, with specific linkages enriched in particular locations during cellular stress. During oxidative stress induced by sodium arsenite, K63-linked polyubiquitin chains accumulate primarily in non-cytosolic compartments, suggesting specialized roles in organelle-specific stress response pathways [45]. This localized ubiquitin signaling is regulated by the ATPase valosin-containing protein (VCP/p97) and its adaptor NPLOC4, which process K63-ubiquitinated substrates in a spatially restricted manner [45].
The development of subcellular ubiquitin proteomics has expanded the known landscape of ubiquitinated proteins by 2.5-fold, identifying 2,494 proteins and 10,157 ubiquitination sites that are compartmentalized during stress conditions [45]. These findings highlight the importance of moving beyond whole-cell ubiquitinomics to understand the spatial regulation of ubiquitin signaling, particularly for atypical chains that may exert their functions in specific subcellular locales.
Figure 2: Functional Landscape of Atypical Ubiquitin Linkages
Applications in Targeted Protein Degradation and Drug Discovery
The expanding knowledge of ubiquitin signaling, particularly regarding linkage specificity, has profound implications for drug discovery, especially in the field of targeted protein degradation (TPD). Mass spectrometry-based ubiquitinomics has become an essential tool for characterizing the mechanism of action of molecular glue degraders (MGDs) and proteolysis-targeting chimeras (PROTACs). For example, high-throughput DIA-MS profiling has been used to map the neosubstrate landscape of cereblon (CRBN)-recruiting ligands, leading to the discovery of novel degraders targeting previously uncharacterized neosubstrates including KDM4B, G3BP2, and VCL [46].
These ubiquitinomics approaches enable researchers to simultaneously monitor ubiquitination changes and consequent protein abundance shifts across the proteome, distinguishing degradative ubiquitination events from non-degradative regulatory ubiquitination. This capability is particularly valuable for understanding the specificity of TPD compounds and for identifying potential off-target effects. Recent studies have demonstrated that only a fraction of proteins showing increased ubiquitination following DUB inhibition or molecular glue treatment are ultimately degraded, highlighting the importance of integrated proteome and ubiquitinome profiling for accurate mechanism of action studies [43].
Mass spectrometry-based ubiquitinomics has evolved from a specialized proteomic application to a mature platform for comprehensive analysis of ubiquitin signaling. The ongoing development of improved sample preparation methods, advanced DIA acquisition strategies, and sophisticated computational tools has dramatically expanded our ability to detect and quantify the ubiquitinome, including the historically challenging atypical K6, K27, and K29 linkages. These technical advances are revealing the remarkable complexity of the ubiquitin code and its spatial regulation within cells.
Future developments in the field will likely focus on improving the throughput and sensitivity of ubiquitinomics workflows, enhancing our ability to characterize complex ubiquitin chain architectures including branched polymers, and developing better computational methods for predicting linkage type from mass spectrometry data. Additionally, the integration of ubiquitinomics with other 'omics datasets will provide a more systems-level understanding of how ubiquitin signaling coordinates cellular processes. As these methods continue to mature, they will undoubtedly uncover new biological functions for atypical ubiquitin chains and accelerate the development of therapeutics that target the ubiquitin-proteasome system.
The ubiquitin code, one of the most complex post-translational signaling systems in eukaryotes, encompasses eight distinct linkage types through which ubiquitin molecules can form polyubiquitin chains. While the functions of K48- and K63-linked chains in proteasomal degradation and signal transduction, respectively, are well-established, the physiological roles of the "atypical" chains (K6, K27, K29) remain largely enigmatic [47]. Genetic interaction analyses using ubiquitin mutants provide a powerful, unbiased approach to uncover pathways regulated by these atypical ubiquitin chain linkages. This methodology enables systematic mapping of functional relationships between ubiquitin linkage types and specific biological processes, bypassing the limitations of biochemical methods in studying rare chain types and revealing unexpected connections in the ubiquitin-proteasome system [48] [49].
The strategic importance of elucidating these pathways has intensified with the growing recognition that atypical ubiquitin chains contribute to vital processes including DNA damage repair, mitophagy, and immune responses [48] [24]. Furthermore, mutations in ubiquitin system components and disruptions in ubiquitin signaling are implicated in numerous pathologies, making this area crucial for therapeutic development [7] [50]. This technical guide outlines the core methodologies for conducting genetic screens with ubiquitin mutants, focusing specifically on applications for characterizing the functions of K6, K27, and K29 chain linkages.
Core Principles: Ubiquitin Chain Diversity and Genetic Screening
The Ubiquitin Code and Atypical Chain Functions
Ubiquitin contains seven lysine residues (K6, K11, K27, K29, K33, K48, K63) and an N-terminal methionine (M1), each capable of forming structurally and functionally distinct polyubiquitin chains [47]. The atypical chains (K6, K27, K29) are relatively rare but play critical roles in specific cellular contexts:
- K6-linked chains: Generated by BRCA1-BARD1 E3 ligase in DNA damage response and by Parkin in mitophagy, functioning predominantly in a proteolysis-independent manner [48] [24].
- K27-linked chains: Also synthesized by Parkin during mitophagy and implicated in immune signaling pathways [48] [49].
- K29-linked chains: Regulate mRNA stability through modification of HuR, an mRNA-binding protein, facilitating its release from specific mRNAs via recognition by UBXD8, a p97 ATPase adaptor [48] [24].
The functional characterization of these linkages has been hampered by their low abundance, technical limitations in detection methods, and redundancy within the ubiquitin system [24] [49].
Genetic Interaction Screening Fundamentals
Genetic interaction analysis examines the phenotypic consequences of combining mutations, revealing functional relationships between genes [49]. A synthetic genetic interaction occurs when two mutations combine to produce a more severe phenotype than expected from their individual effects, indicating the genes likely function in parallel pathways or processes. In contrast, epistasis describes a situation where the double mutant phenotype resembles one of the single mutants, suggesting the genes operate in the same linear pathway [48].
When applied to ubiquitin linkage studies, this approach involves systematically combining mutations that eliminate specific ubiquitin acceptor lysines with deletions of non-essential genes across the genome. The resulting growth phenotypes of double mutant strains reveal which cellular processes become essential when specific ubiquitin linkages are compromised [24] [49].
Methodology: Implementing Ubiquitin Mutant Screens
Strain Engineering and Ubiquitin Mutant Design
A critical first step involves engineering yeast strains expressing ubiquitin mutants as the sole source of ubiquitin. Since ubiquitin is encoded by four loci in Saccharomyces cerevisiae (UBI1-4), this requires modifying all four loci to express the desired ubiquitin variant while preserving essential functions of ubiquitin fusion proteins [48] [49].
Key considerations for strain design:
- Lysine-to-arginine (K-to-R) mutations: Substitute specific acceptor lysines with arginine to prevent chain formation through that site while preserving ubiquitin's charge and structure [24].
- Single-lysine ubiquitin variants: For gain-of-function studies, create ubiquitin mutants where all lysines except one are mutated to arginine, forcing all chain formation through a single linkage type [49].
- Viability controls: Since K48-linked chains are essential for viability, strains studying K48R mutations must retain some wild-type ubiquitin expression (typically 20%) [24].
- Expression level validation: Ensure mutant ubiquitin strains express ubiquitin at levels comparable to wild-type strains to avoid confounding effects of reduced ubiquitin availability [24].
Table 1: Essential Research Reagents for Ubiquitin Genetic Screens
| Research Reagent | Function and Application | Key Features and Considerations |
|---|---|---|
| K-to-R Ubiquitin Mutants | Eliminate specific ubiquitin chain linkages | Arg substitution preserves charge; K48R requires WT ubiquitin co-expression |
| Single-Lysine Ubiquitin Variants | Force chain formation through single linkage type | Conditional expression often necessary due to lethality |
| SK1 Yeast Strain Background | High-efficiency sporulation essential for multi-locus screening | ~92% sporulation efficiency vs. ~12% in S288C |
| Modified UBI1 Locus (Rpl40A expressed separately) | Reduces selection markers needed in cross | Uses constitutive GPD promoter; decreases number of selected loci |
| Haploid Selection Markers | Select for desired haploid progeny during SGA | Typically 4-6 selectable markers required for ubiquitin SGA |
Modified Synthetic Genetic Array (SGA) Protocol
Conventional SGA methods require optimization for ubiquitin studies due to the need to manipulate multiple ubiquitin loci simultaneously. The following protocol outlines key modifications for ubiquitin linkage screens [48] [49]:
A. Strain Preparation (0-2 days)
- Generate query strains expressing ubiquitin mutants in the SK1 background, with all four ubiquitin loci modified to express the desired ubiquitin variant.
- Maintain the gene deletion library (array strains) in the same SK1 background with equivalent genetic modifications (e.g., UBI1 locus replacement).
B. Mating and Diploid Selection (2-5 days)
- Cross query strains with arrayed gene deletion library by replica pinning onto rich medium (YEPD).
- Incubate overnight to allow mating.
- Transfer to medium selecting for diploids (e.g., lacking amino acids corresponding to both parental markers).
- Incubate for 2 days to select for successful diploids.
C. Sporulation and Haploid Selection (5-12 days)
- Transfer diploids to nitrogen-deficient sporulation medium.
- Incubate for 5-7 days to induce sporulation.
- Transfer to medium selecting for haploid progeny containing both the ubiquitin mutations and gene deletion of interest.
- Due to SK1's high sporulation efficiency, omit one haploid selection step (e.g., lyp1Δ selection) to improve efficiency.
D. Phenotypic Analysis (12-19 days)
- Measure colony sizes of double mutant strains using high-resolution scanning.
- Normalize growth measurements to control strains.
- Calculate genetic interaction scores (S-scores) to identify significant interactions.
Diagram 1: Ubiquitin SGA screening workflow
Data Analysis and Genetic Interaction Scoring
Genetic interaction scoring methodology:
- Colony size normalization: Normalize double mutant colony sizes to account for systematic technical variations.
- S-score calculation: Compute S-scores using established SGA protocols, where scores are centered around zero, with approximately 53% negative and 47% positive values in typical ubiquitin SGA datasets [49].
- Threshold determination: Set appropriate thresholds for significant genetic interactions based on score distributions (typically |S-score| > 2-3).
- Clustering analysis: Group genes with similar genetic interaction profiles to identify functional modules dependent on specific ubiquitin linkages.
Quality control measures:
- Include control strains with known genetic interactions to validate screening methodology.
- Perform replicate experiments to assess reproducibility.
- Exclude ubiquitin mutants with extreme hypersensitivity to selective agents (e.g., K63R mutants show canavanine hypersensitivity) [24].
Application to Atypical Ubiquitin Chain Research
Interpreting Genetic Interactions for K6, K27, and K29 Linkages
For atypical ubiquitin chains, genetic interactions reveal pathways that become essential when specific linkages are compromised. The interpretation framework includes:
Negative genetic interactions (synthetic sickness/lethality) indicate parallel pathways or redundant functions. For example, if a K29R ubiquitin mutant shows synthetic lethality with deletions in DNA repair genes, this suggests K29-linked chains function redundantly with these genes in maintaining genomic stability.
Positive genetic interactions (alleviating interactions) suggest the genes function in the same pathway. If a mutation in a specific E3 ligase ameliorates the growth defect of a K27R mutant, this might indicate that ligase generates K27-linked chains that are detrimental in certain contexts.
Cluster analysis identifies groups of genes with similar genetic interaction profiles to the ubiquitin mutant, revealing functional networks dependent on that specific ubiquitin linkage type [49].
Validation of Candidate Interactions
Initial genetic interactions require validation through independent methods:
A. Low-throughput validation
- Generate fresh haploid double mutants using standard yeast genetic techniques.
- Perform serial dilution spot assays on appropriate media to quantitatively assess growth defects.
- Compare growth rates of single versus double mutants in liquid culture.
B. Molecular mechanism investigation
- Assess substrate ubiquitination in ubiquitin mutant strains using linkage-specific tools (e.g., TUBEs, linkage-specific antibodies) [7] [51].
- Measure protein turnover rates of pathway components in ubiquitin mutant backgrounds.
- Analyze pathway activity through transcriptional reporters or biochemical assays.
Table 2: Example Genetic Interactions for Atypical Ubiquitin Chains
| Ubiquitin Mutant | Interacting Gene/Pathway | Interaction Type | Proposed Functional Connection |
|---|---|---|---|
| K11R | APC/C subunit (Apc9) | Negative | K11-linkages work with APC/C for cell cycle regulation |
| K11R | Threonine biosynthetic genes | Negative | K11-linkages important for amino acid import |
| K29R | mRNA binding proteins | Negative | K29-linkages regulate mRNA stability via HuR |
| K6R | DNA damage response genes | Negative | K6-linkages function in BRCA1-mediated DNA repair |
| K27R | Mitophagy regulators | Negative | K27-linkages participate in Parkin-mediated mitophagy |
Technical Considerations and Advanced Applications
Optimization for Specific Ubiquitin Linkages
The methodology requires specific adaptations for studying atypical chains:
Enhancing sensitivity for rare linkages:
- Use sensitized backgrounds (e.g., additional stress conditions) to reveal phenotypes for rarely used linkages.
- Combine multiple atypical chain mutations (e.g., K27R/K29R double mutant) to address potential redundancy.
- Employ chemical inhibitors of parallel pathways to unmask dependencies on specific ubiquitin linkages.
Experimental controls:
- Include strains with low wild-type ubiquitin expression to control for general ubiquitin level effects.
- Use linkage-specific ubiquitin binding domains to verify the biochemical consequences of ubiquitin mutations.
- Monitor chain type abundance in mutants to confirm expected linkage reduction.
Integration with Biochemical and Proteomic Methods
Genetic interaction data gains greater power when integrated with complementary approaches:
Mass spectrometry-based ubiquitin proteomics:
- Use AQUA (Absolute QUAntification) with labeled ubiquitin peptide standards to quantify linkage abundance in mutants [47] [51].
- Employ diGly capture proteomics to identify changes in global ubiquitination patterns in ubiquitin mutant strains.
Biochemical validation:
- Apply Tandem Ubiquitin Binding Entities (TUBEs) with linkage specificity to monitor changes in substrate ubiquitination in different mutant backgrounds [7] [52].
- Utilize linkage-specific deubiquitinases (DUBs) as analytical tools to verify chain types present on pathway components [47].
Diagram 2: Multi-modal data integration
Genetic interaction analyses using ubiquitin mutants provide a powerful, systematic approach to uncover pathways regulated by atypical ubiquitin chain linkages. The methodology outlined here enables researchers to move beyond the well-characterized K48 and K63 linkages and explore the functional landscape of K6, K27, and K29 chains in specific biological contexts. By integrating these genetic approaches with biochemical validation methods, researchers can construct comprehensive models of how atypical ubiquitin linkages contribute to cellular homeostasis and human disease, ultimately facilitating targeted therapeutic interventions that exploit specific aspects of the ubiquitin code.
The post-translational modification of proteins with polyubiquitin chains is a fundamental regulatory mechanism in eukaryotic cells. While K48- and K63-linked chains have been extensively characterized, the so-called "atypical" ubiquitin linkages—K6, K27, and K29—have emerged as crucial regulators in specialized cellular processes. These atypical linkages confer unique structural properties and functional outcomes that distinguish them from their canonical counterparts. K27-linked ubiquitination has been implicated in mitochondrial quality control and innate immunity regulation, often exhibiting remarkable resistance to deubiquitinase (DUB) activity [14]. K29-linked chains function in transcriptional regulation during the unfolded protein response (UPR) and Wnt signaling pathways [23]. K6-linked chains are associated with DNA damage repair processes [15]. Understanding the distinct biological functions of these linkages requires elucidation of their three-dimensional structures and dynamics, which often exist as conformational ensembles rather than single, static structures.
The inherent flexibility and dynamic nature of atypical ubiquitin chains presents a substantial challenge for classical high-resolution structural techniques like X-ray crystallography. This technical guide details how integrated approaches using Nuclear Magnetic Resonance (NMR) spectroscopy and Small Angle Neutron Scattering (SANS) provide complementary solutions to characterize the conformation and dynamics of K6, K27, and K29-linked ubiquitin chains, enabling researchers to connect structural insights to biological function.
Technical Foundations: Principles of NMR and SANS
Nuclear Magnetic Resonance (NMR) Spectroscopy
NMR spectroscopy exploits the magnetic properties of atomic nuclei to obtain atomic-resolution information on protein structure and dynamics in solution. For ubiquitin chains, NMR provides residue-specific data across multiple timescales, from picosecond bond vibrations to second-scale conformational exchanges.
Key observables in NMR studies of atypical ubiquitin chains include:
- Chemical Shifts: Sensitive indicators of local electronic environment used to identify binding interfaces and conformational changes through Chemical Shift Perturbation (CSP) analysis [53] [14].
- Residual Dipolar Couplings (RDCs): Provide long-range orientational restraints for determining domain arrangements within flexible chains, especially valuable for systems with sparse distance restraints [53].
- Relaxation Parameters: Characterize molecular dynamics across picosecond-to-nanosecond and microsecond-to-millisecond timescales, quantifying conformational flexibility [54] [55].
- Paramagnetic Effects: Paramagnetic relaxation enhancement (PRE) from spin labels provides long-range distance restraints up to 20-25 Å, crucial for mapping transient interdomain contacts [53].
Advanced NMR methods have been developed specifically for challenging systems like ubiquitin chains. The ultra-selective 1H-15N NMR method (SNIPER) enables high-quality measurement of individual 15N spin-relaxation constants even when 15N resonances are heavily congested, as commonly encountered in disordered protein regions [54]. 19F NMR with parallel incorporation of multiple fluorinated aromatic amino acids (FAAAs) offers enhanced sensitivity for studying protein conformation and interactions [56].
Small Angle Neutron Scattering (SANS)
SANS measures the elastic scattering of neutrons at small angles, providing low-resolution information about the overall shape, size, and organization of macromolecules in solution. Unlike NMR, SANS has no inherent size limitations, making it particularly valuable for studying large complexes or polymers.
The key advantage of SANS for ubiquitin chain research lies in contrast variation through selective deuteration. By preparing samples with specific deuterated components and manipulating the D₂O concentration in the solvent, researchers can effectively "match out" the scattering contribution from selected parts of a complex, enabling individual subunits to be highlighted within a larger assembly [53] [57]. For a ternary protein-protein-RNA complex, for instance, SANS with subunit-selectively deuterated samples can reveal the positioning of individual components within the overall structure [53].
Recent methodological advances have significantly enhanced SANS capabilities:
- High-Throughput SANS: Automated size-exclusion-chromatography coupled with SANS at synchrotron beamlines enables rapid sample characterization [53].
- Accelerated Data Acquisition: Novel algorithms optimize neutron counting by estimating the minimum counts required for reliable parameter estimation, dramatically reducing measurement time [58].
- Time-Resolved SANS: Enables monitoring of structural changes in real-time under various perturbations [57] [58].
Integrated Methodologies for Atypical Ubiquitin Chains
Sample Preparation and Labeling Strategies
The study of atypical ubiquitin chains begins with the production of well-defined, homogeneously linked chains. This is particularly challenging for K6, K27, and K29 linkages due to the limited availability of linkage-specific enzymes.
Table 1: Key Research Reagent Solutions for Atypical Ubiquitin Chain Studies
| Reagent/Technique | Function | Application Examples |
|---|---|---|
| Non-enzymatic Ubiquitin Chain Assembly [14] | Chemical synthesis of native isopeptide linkages using mutually orthogonal removable amine-protecting groups (Alloc and Boc) | Production of fully natural K6-, K27-, and K29-Ub₂s with native isopeptide linkages, free of mutations |
| Selective Deuteration [53] [57] | Partial or complete replacement of ¹H with ²H to reduce relaxation pathways in NMR or enable contrast matching in SANS | Deuteration of specific subunits in complexes for SANS contrast variation; perdeuteration for TROSY-based NMR of large complexes |
| Isotope Labeling [53] | Incorporation of ¹⁵N, ¹³C for NMR detection; specific protonation of methyl groups in deuterated background | ¹⁵N-labeling for 1H-15N correlation experiments; methyl labeling of Ile, Leu, Val for NMR studies of high molecular weight complexes |
| Paramagnetic Tags [53] | Covalent attachment of nitroxide spin labels or lanthanide binding tags (LBTs) to cysteine residues | Generation of long-range distance restraints (up to 25 Å) through paramagnetic relaxation enhancement (PRE) |
| Fluorinated Aromatic Amino Acids [56] | Incorporation of 3-fluorotyrosine, 4-fluorophenylalanine, or 5-fluorotryptophan for ¹⁹F NMR | Enhanced sensitivity for studying conformational changes and interactions in complex ubiquitin chains |
For NMR studies, atypical ubiquitin chains require specific isotopic labeling patterns. Uniform ¹⁵N-labeling enables basic fingerprinting through 1H-15N correlation experiments, while ¹³C/¹⁵N-labeling facilitates backbone assignment. For larger complexes, selective methyl labeling (of isoleucine, leucine, and valine) in a deuterated background significantly improves sensitivity [53]. For SANS experiments, strategic deuteration is crucial. Preparing samples with perdeuterated proximal ubiquitin and protonated distal ubiquitin (or vice versa) enables precise determination of inter-domain arrangements through contrast matching [53].
Experimental Workflows and Data Acquisition
The integrated NMR/SANS approach follows a logical workflow that progresses from sample validation to high-resolution structural and dynamic characterization.
Diagram 1: Integrated NMR-SANS Workflow for Atypical Ubiquitin Chains
NMR Protocol for Atypical Ubiquitin Chain Characterization
1. Basic Characterization and Binding Interface Mapping
- Experiment: 1H-15N Heteronuclear Single Quantum Coherence (HSQC)
- Parameters: Typically acquired with 1024 (¹H) × 256 (¹⁵N) complex points, spectral widths of 14 ppm (¹H) and 30 ppm (¹⁵N) [14]
- Analysis: Chemical Shift Perturbation (CSP) calculated using the formula: CSP = √(ΔδH² + (0.2×ΔδN)²), where ΔδH and ΔδN are chemical shift changes in ¹H and ¹⁵N dimensions, respectively [14]
- Application: For K27-Ub₂, CSP analysis revealed widespread perturbations in the proximal ubiquitin but minimal changes in the distal ubiquitin, suggesting absence of stable interdomain contacts [14]
2. Dynamics Measurements
- Fast Dynamics (ps-ns): ¹⁵N R₁, R₂, and {¹H}-¹⁵N NOE measurements
- Slow Dynamics (μs-ms): CPMG relaxation dispersion, ZZ-exchange spectroscopy
- Application: The SNIPER pulse sequence enables measurement of individual ¹⁵N spin-relaxation constants even for highly congested spectra of low-complexity regions, as demonstrated for huntingtin polyglutamine stretches [54]
3. Long-Range Restraints
- Residual Dipolar Couplings (RDCs): Measured in weakly aligning media (phages, gels)
- Paramagnetic Relaxation Enhancement (PRE): Using cysteine-conjugated nitroxide spin labels (e.g., MTSL)
SANS Protocol for Atypical Ubiquitin Chain Characterization
1. Basic Data Collection
- Instrument Configuration: Typically utilizes two detector positions to cover a wide q-range (e.g., 15 m for 0.0037-0.05 Å⁻¹ and 2 m for 0.03-0.43 Å⁻¹) [58]
- Sample Preparation: Ubiquitin chains in appropriate buffers (e.g., phosphate buffer, pH 7.0-7.6), with matching D₂O/H₂O ratios for contrast variation
- Measurement Time: Typically 10 minutes to several hours per condition, depending on scattering power [58]
2. Contrast Variation SANS
- Sample Design: Prepare ubiquitin chains with specific deuterated components
- D₂O Series: Collect data at multiple D₂O concentrations (e.g., 0%, 42%, 70%, 100%)
- Analysis: Using the scattering length density (SLD) matching points (42% D₂O for protonated protein, 70% D₂O for RNA) to highlight specific components [53]
3. Data Processing and Analysis
- Radius of Gyration: From Guinier analysis: I(q) = I(0)exp(-(qRg)²/3) for qRg < 1.3
- Pair Distance Distribution: P(r) function calculation via indirect Fourier transform
- Shape Modeling: Ab initio shape reconstruction using programs like DAMMIF [53]
Data Integration and Computational Modeling
The true power of the integrated approach emerges when NMR and SANS data are combined to generate conformational ensembles that represent the dynamic reality of atypical ubiquitin chains in solution.
Hybrid Modeling Approaches:
- NMR-Driven Docking with SANS Validation: NMR-derived structural models (from CSPs, PREs, RDCs) are validated against experimental SANS data to exclude models inconsistent with the overall shape [53]
- Ensemble Modeling Methods: Computational approaches (e.g., ENSEMBLE, MES) generate a collection of structures that collectively satisfy both atomic-level NMR restraints and low-resolution SANS data [14]
- Bayesian/Maximum Entropy Methods: Derive conformational ensembles that best satisfy all experimental constraints while maximizing conformational entropy
For K27-Ub₂, this integrated approach revealed an extended conformation with high flexibility and absence of stable interdomain contacts, explaining its unique resistance to deubiquitinases [14].
Application to Atypical Ubiquitin Linkages: Key Findings
Structural and Dynamic Properties of K6, K27, and K29 Linkages
The application of integrated NMR/SANS approaches has revealed distinctive structural features for each atypical ubiquitin linkage.
Table 2: Structural and Dynamic Properties of Atypical Ubiquitin Chains from NMR and SANS Studies
| Linkage | Overall Architecture | Dynamic Properties | Functional Implications |
|---|---|---|---|
| K6-Linked Ub₂ [14] | Compact conformation with non-covalent interdomain contacts involving the hydrophobic patch (L8, I44, V70) | Restricted flexibility compared to extended linkages | DNA damage repair functions; recognized by BRCA1-BARD1 complex |
| K27-Linked Ub₂ [14] | Extended conformation with no stable interdomain contacts | High flexibility and conformational heterogeneity | Resistant to most deubiquitinases; regulates mitochondrial autophagy and innate immunity |
| K29-Linked Ub₂ [14] [23] | Weak or transient interdomain contacts | Moderate flexibility | Transcriptional regulation during UPR; Wnt signaling; mRNA stability control |
Case Study: K27-Linked Diubiquitin (K27-Ub₂)
The comprehensive analysis of K27-Ub₂ exemplifies the power of integrated NMR/SANS approaches. Key findings include:
Unique Deubiquitinase Resistance:
- K27-Ub₂ resists cleavage by multiple DUB families including USP2, USP5 (IsoT), and Ubp6 [14]
- This distinguishes K27 from all other linkages and suggests unique structural features
Distinct Structural Features from NMR:
- Minimal CSPs in distal ubiquitin indicate absence of stable noncovalent interdomain contacts [14]
- Widespread CSPs in proximal ubiquitin centered around K27 and extending to adjacent regions
- Dynamic properties indicate high flexibility and conformational heterogeneity
SANS Validation:
- Confirms extended overall shape inconsistent with compact conformations
- Ensemble modeling reconciles NMR and SANS data, revealing a dynamic conformational landscape
Functional Correlation:
- The extended, flexible conformation may prevent optimal engagement with DUB active sites
- Recognition by proteasomal shuttle protein hHR23a UBA2 domain suggests functional versatility [14]
Biological Context and Therapeutic Implications
The structural insights gained from NMR and SANS studies directly illuminate the biological functions of atypical ubiquitin chains:
K27 Linkages in Disease:
- K27-polyubiquitination of Miro1 regulates mitochondrial trafficking and damage response [14]
- Implicated in regulation of innate immunity pathways [14]
K29 Linkages in Transcriptional Regulation:
- Chromatin landscape analysis reveals K29 ubiquitin chains enriched at promoter regions with transcriptional activation marks (H3K4me3, H3K27ac) [23]
- During unfolded protein response, K29 ubiquitination of cohesin complex (SMC1A, SMC3) regulates transcription of cell proliferation-related genes [23]
Therapeutic Targeting Opportunities:
- Unique structural features of atypical linkages offer potential for selective therapeutic targeting
- DUB resistance of K27 chains suggests possible strategies for modulating ubiquitin signaling
- The conformational ensembles provide templates for structure-based drug design
The integration of NMR spectroscopy and Small Angle Neutron Scattering provides a powerful methodological framework for elucidating the structure and dynamics of atypical ubiquitin chains. This approach bridges the resolution gap between atomic-level detail and overall shape characterization, enabling comprehensive understanding of these dynamic systems in solution. For the atypical linkages K6, K27, and K29, this has revealed distinctive structural features that explain their unique functional properties and cellular roles.
Future advancements in this field will likely include:
- Increased Application of 19F NMR: Utilizing parallel incorporation of multiple fluorinated aromatic amino acids for enhanced sensitivity in studying conformational changes and interactions [56]
- Advanced SANS Methodologies: Implementation of accelerated data acquisition algorithms and machine learning approaches to optimize beam time usage and data analysis [58]
- Time-Resolved Studies: Application of stopped-flow SANS and real-time NMR to capture ubiquitin chain dynamics during enzymatic assembly and disassembly
- Cellular Context Applications: Developing approaches to extend these structural insights into more complex cellular environments
The continued refinement and application of integrated NMR/SANS approaches will undoubtedly yield further insights into the structural biology of atypical ubiquitin chains, advancing both fundamental understanding and therapeutic targeting of these important signaling molecules.
Activity-Based Probes for Deubiquitinases (DUBs) with Linkage Specificity
The ubiquitin code, a complex post-translational language, governs virtually all eukaryotic cellular processes. This complexity extends beyond the well-characterized K48- and K63-linked ubiquitin chains to encompass atypical linkages (K6, K11, K27, K29, K33), whose specific functions and regulation remain inadequately decoded [8] [36]. Deubiquitinases (DUBs) are crucial editors of this code, and their linkage-specific activities are paramount for maintaining cellular homeostasis. Traditional methods often fall short in capturing the dynamic activity of DUBs toward specific chain types. Activity-Based Probes (ABPs) have thus emerged as indispensable chemical tools that transform DUBs from mere subjects of study into direct reporters of their own catalytic functions and specificities [59] [36]. These probes facilitate the functional annotation of DUBs, particularly for the least understood atypical linkages, enabling the identification of novel DUB activities, mechanistic dissection of their catalytic pathways, and screening for selective inhibitors with therapeutic potential. This technical guide details the composition, application, and experimental protocols for using ABPs to profile linkage-specific DUB activities, with a special emphasis on the K6, K27, and K29 chain types central to advanced ubiquitin research.
The Molecular Architecture of DUB ABPs
Activity-Based Probes for DUBs are engineered to covalently trap active enzymes, thereby providing a snapshot of their catalytic state and specificity. The design of these probes is modular, typically consisting of three core elements:
- Ubiquitin or Ubiquitin Chain Scaffold: This serves as the recognition element. While monoubiquitin is used for general DUB profiling, linkage-defined polyubiquitin chains are critical for determining linkage specificity. For instance, K63-linked tetraubiquitin chains were essential for identifying USP53 and USP54 as K63-specific DUBs [60]. To study atypical linkages, homotypic K6-, K27-, or K29-linked chains are required.
- Warhead Group: This is a reactive electrophile that forms a covalent bond with the catalytic cysteine residue in the DUB's active site. Common warheads include:
- Propargylamide (PA): Forms a vinyl thioether adduct with the catalytic cysteine, enabling enrichment and identification of active DUBs [60].
- Michael Acceptors: Warheads such as vinyl sulfone or dehydroalanine can also act as potent electrophiles for covalent modification.
- Detection/Affinity Handle: This moiety allows for the purification, detection, or enrichment of the DUB-probe complex. Frequently used handles include:
- Hemagglutinin (HA) tag: For immunodetection.
- Biotin: For streptavidin-based enrichment.
- Fluorescent tags: For in-gel visualization.
Table 1: Common Warheads and Handles in DUB ABPs
| Component Type | Specific Example | Key Feature | Primary Application |
|---|---|---|---|
| Warhead | Propargylamide (PA) | Forms stable vinyl thioether | Active-site profiling & enrichment [60] |
| Warhead | Vinyl Sulfone | Michael acceptor | General activity-based profiling |
| Affinity Handle | HA tag | High-affinity antibodies | Immunodetection and immunoprecipitation |
| Affinity Handle | Biotin | Strong streptavidin binding | Streptavidin pull-down and mass spectrometry |
| Report Handle | Rhodamine 110 (RhoG) | Fluorogenic | Real-time kinetic assays [60] |
| Report Handle | TAMRA | Fluorescent | Gel-based activity monitoring [60] |
A critical application of these probes is the discovery and validation of DUBs with unique linkage preferences. A seminal example is the recent reannotation of USP53 and USP54. Previously considered catalytically inactive pseudoenzymes, they were revealed as highly specific K63-linkage-directed DUBs through the use of a HA-Ub-PA probe and linkage-defined tetraubiquitin cleavage assays [60]. This finding, driven by ABP technology, underscores the potential for discovering novel DUB activities against other linkage types, including the atypical chains.
Diagram 1: ABP mechanism for covalent DUB labeling.
Methodologies for Profiling Linkage-Specific DUB Activity
Direct DUB Labeling and Enrichment with ABPs
This protocol uses ABPs to covalently label active DUBs in complex protein mixtures, allowing for their subsequent enrichment and identification.
- Sample Preparation: Prepare cell lysates in a non-denaturing buffer. For a controlled assessment, use purified recombinant DUB catalytic domains (e.g., USP53, USP54) [60].
- Probe Incubation: Incubate the sample with the desired ABP (e.g., HA-Ubiquitin-PA). A typical reaction uses 1–5 µM probe for 30–60 minutes at a physiological temperature (e.g., 30°C) [60].
- Enrichment: For HA-tagged probes, use anti-HA antibody-conjugated beads. For biotinylated probes, use streptavidin-coated beads. Incubate the probe-labeled lysate with the beads for 1–2 hours at 4°C.
- Washing and Elution: Wash beads stringently to remove non-specifically bound proteins. Elute bound proteins by boiling in SDS-PAGE loading buffer.
- Analysis: Analyze the eluates by immunoblotting for specific DUBs or by mass spectrometry for global DUB identification.
In Vitro Deubiquitination Assays with Defined Ubiquitin Chains
This assay directly measures a DUB's catalytic activity and linkage specificity by monitoring the cleavage of defined ubiquitin substrates.
- Substrate Preparation: Source linkage-defined polyubiquitin chains. These can be produced enzymatically, by chemical synthesis, or via thioether-mediated ligation, which is particularly useful for generating atypical linkages like K27 and K29 [61].
- Reaction Setup: Combine the DUB (e.g., purified catalytic domain of USP53) with the ubiquitin chain substrate (e.g., K63-linked tetraubiquitin) in a suitable reaction buffer. A standard reaction might contain 100–500 nM DUB and 1–5 µM ubiquitin chain [60].
- Incubation and Time-Course Sampling: Incubate at a relevant temperature (e.g., 30°C or 37°C). Remove aliquots at various time points (e.g., 0, 5, 15, 30, 60 minutes) and quench immediately by adding SDS-PAGE loading buffer.
- Product Analysis: Resolve the quenched samples by SDS-PAGE. Visualize the cleavage products using ubiquitin-specific antibodies or by staining (e.g., Coomassie, silver stain). The disappearance of the input chain and the appearance of shorter chains or free ubiquitin indicate DUB activity.
Table 2: Analysis of USP53/USP54 Cleavage of K63-linked Tetraubiquitin
| Time Point (min) | Substrate (TetraUb) | Primary Product (DiUb) | Final Product (MonoUb) | Interpretation |
|---|---|---|---|---|
| 0 | 100% | 0% | 0% | No activity |
| 5 | ~60% | ~40% | 0% | Rapid endo-cleavage |
| 30 | ~10% | ~70% | ~20% | Processive cleavage |
| 60 | 0% | ~30% | ~70% | Near-complete substrate turnover |
Fluorogenic DUB Activity Assays
For high-throughput screening and kinetic analysis, fluorogenic ubiquitin substrates are invaluable.
- Probe Selection: Use C-terminally derivatized ubiquitin conjugated to a fluorophore like Rhodamine 110 (Ub-RhoG). The quenched probe fluoresces upon DUB-mediated cleavage [60].
- Kinetic Measurement: In a microplate reader, mix the DUB with Ub-RhoG (typically 50–200 nM probe). Monitor the increase in fluorescence (excitation/~485 nm, emission/~525 nm) over time.
- Data Analysis: Calculate initial reaction velocities to determine catalytic efficiency. This assay is ideal for inhibitor screening and comparative analysis of DUB activity against different probe variants.
The Scientist's Toolkit: Essential Research Reagents
Success in profiling linkage-specific DUBs relies on a suite of specialized reagents and tools.
Table 3: Key Reagent Solutions for DUB and Atypical Linkage Research
| Reagent/Tool | Function | Example in Research |
|---|---|---|
| Linkage-Defined Ubiquitin Chains | Substrates for determining DUB linkage specificity in vitro. | K63-linked tetraubiquitin revealed USP53/USP54 specificity [60]. |
| ABPs with Atypical Linkages | Profiling DUBs active on rare chains (K6, K27, K29). | Critical for expanding research beyond K48/K63 [36]. |
| Linkage-Specific Antibodies/Affimers | Detecting endogenous levels of specific ubiquitin linkages. | K29/K48-bispecific antibody helps study branched chains [36]. |
| Ubiquitin Replacement Cell Lines | Studying cellular function of specific linkages. | Cell lines expressing only K29R mutant Ub reveal K29 linkage roles [62]. |
| Thioether Ligation Kit | Chemically generating homogeneous ubiquitinated proteins. | Protocol for constructing probes like di-Ub-PCNA [61]. |
Diagram 2: Experimental workflow for linkage-specific DUB characterization.
Case Study: Connecting K29-Linked Ubiquitination to Chromatin Regulation
Research into atypical linkages is revealing their critical, non-redundant roles. A prime example is K29-linked ubiquitination. A comprehensive profiling of ubiquitin linkage functions using ubiquitin-replacement cell lines identified a strong association between K29-linked chains and chromosome biology [62]. This study identified the histone methyltransferase SUV39H1—a key enzyme responsible for depositing the repressive H3K9me3 mark—as a major substrate for K29-linked ubiquitylation. The E3 ligase TRIP12 was identified as the enzyme catalyzing K29-linked ubiquitination of SUV39H1, targeting it for proteasomal degradation [62]. This K29-linked degradation signal is reversed by the DUB TRABID, which contains specific binding domains for K29-linked chains [62]. This precise regulation is essential for maintaining H3K9me3 homeostasis and epigenome integrity. Disrupting K29-linked ubiquitylation of SUV39H1 by ablating the linkage or depleting TRIP12 led to SUV39H1 stabilization and deregulation of heterochromatin marks [62]. This case highlights a dedicated K29-linked ubiquitination pathway and underscores the urgent need for ABPs that can target the DUBs, like TRABID, which edit this specific code, opening new avenues for drug discovery in epigenetics.
Navigating Experimental Challenges: Overcoming Obstacles in Atypical Ubiquitin Research
The study of atypical ubiquitin chains, particularly those linked through K6, K27, and K29 residues, represents a frontier in understanding sophisticated post-translational regulation of cellular processes. Unlike the well-characterized K48 and K63-linked chains, these atypical linkages often exist at extremely low cellular abundances while exerting profound biological influence. Their investigation is plagued by substantial technical challenges, as high-abundance proteins can obscure detection and accurate quantification of these rare modifications. The dynamic range of protein concentrations in biological samples can span over 10 orders of magnitude, with low-abundance targets like atypical ubiquitin chains frequently sitting below confident detection thresholds using standard proteomic approaches. This technical whitepaper provides a comprehensive guide to advanced methodologies that enable researchers to overcome these limitations, with specific application to the functional characterization of K6, K27, and K29-linked ubiquitin chains in both basic research and drug discovery contexts.
The Biological Significance of Low-Abundance Atypical Ubiquitin Chains
Atypical ubiquitin chains constitute a sophisticated regulatory layer controlling vital cellular processes. While historically challenging to study due to their low abundance and technical limitations, recent advances have revealed their essential functions in antiviral immunity, cell cycle regulation, and protein degradation. K27-linked chains have emerged as particularly important in immune signaling, where they mediate a complex balance between activation and inhibition of innate immune pathways. For example, TRIM23-mediated K27-linked ubiquitination of NEMO activates both NF-κB and IRF3 signaling pathways, while K27/K29-branched chains on MAVS orchestrate its autophagy-mediated degradation to restrict type I interferon responses [8].
The functional diversity of these modifications is extraordinary. K6-linked chains have been implicated in mitophagy and DNA damage response, often working in concert with Parkin E3 ligase to designate damaged mitochondria for destruction [3]. K29-linked chains contribute to proteasomal degradation and mRNA stability regulation, exemplified by their attachment to HuR, an mRNA binding protein [24]. Despite their low abundance, these atypical linkages form a complex "ubiquitin code" that dramatically expands the functional repertoire of ubiquitin signaling, creating a pressing need for sensitive detection and enrichment strategies to decipher their biological roles.
Table 1: Functions of Atypical Ubiquitin Chains in Cellular Regulation
| Ubiquitin Linkage | Biological Functions | E3 Ligases | Cellular Processes |
|---|---|---|---|
| K6-linked | Mitochondrial quality control, DDR | Parkin, HUWE1 | Mitophagy, DNA repair |
| K27-linked | Immune signaling regulation | TRIM23, RNF185 | NF-κB and IRF3 activation, STING signaling |
| K29-linked | Proteasomal targeting, mRNA stability | Ufd4, E6AP | Protein degradation, mRNA regulation |
| K11-linked | Cell cycle progression, ERAD | APC/C, UBE2S | Mitotic regulation, protein degradation |
| K33-linked | Protein trafficking | - | Post-Golgi transport |
Advanced Enrichment Strategies for Low-Abundance Targets
Combinatorial Peptide Ligand Libraries (CPLLs)
Combinatorial Peptide Ligand Library technology represents a powerful approach for addressing the extreme dynamic range of protein concentrations in biological samples. This method employs a mixed bed of affinity sorbents containing millions of unique hexapeptide structures synthesized using combinatorial chemistry. When exposed to complex protein extracts, high-abundance proteins rapidly saturate their binding sites, while low-abundance species continue to concentrate as sample volume increases [63]. The technology effectively reduces concentration disparities from 10-12 orders of magnitude to a more manageable 2-3 orders, bringing previously undetectable targets like atypical ubiquitinated proteins within detection range.
The CPLL workflow involves several critical phases: First, sample loading under physiological conditions to maintain native protein conformations; second, extensive washing to remove non-specifically bound proteins; and finally, elution using conditions that disrupt protein-ligand interactions. An alternative approach involves on-bead digestion of captured proteins followed by LC-MS/MS analysis, which can improve recovery of strongly-bound targets [63]. For ubiquitin chain research, CPLL enrichment can be performed prior to chain-specific immunoprecipitation, dramatically enhancing the detection of low-abundance atypical linkages.
Immunoaffinity-Based Enrichment Methods
Immunoaffinity approaches provide linkage-specific enrichment capabilities essential for atypical ubiquitin chain research. While antibodies against K6, K27, and K29 linkages are less prevalent than those for canonical chains, careful validation using knockout controls can identify reliable reagents. For Western blot applications, four of nine commercially available antibodies tested under knockout validation conditions demonstrated specificity for their targets [64]. However, researchers should exercise caution, as the same study found no antibodies suitable for immunocytochemistry applications, highlighting the importance of rigorous validation for each intended application.
Immunoaffinity methods face particular challenges in ubiquitin research due to the structural similarities between different chain types and the potential for branched chains containing multiple linkage types. Nevertheless, when combined with prior CPLL enrichment, linkage-specific immunoprecipitation can successfully isolate low-abundance atypical chains for downstream analysis. The limited specificity of some commercial antibodies necessitates corroboration using multiple methods to confirm findings.
Chromatographic and Electrophoretic Fractionation
Traditional separation methods including liquid chromatography and electrophoresis remain valuable tools for reducing sample complexity prior to targeted analysis of atypical ubiquitin chains. Multi-dimensional separation approaches significantly expand peak capacity, improving the detection of low-abundance species. These methods are particularly effective when combined with targeted detection methods such as multiple reaction monitoring (MRM) or parallel reaction monitoring (PRM) mass spectrometry.
Chromatographic fractionation can be applied at either the protein or peptide level, with the latter often proving more effective for ubiquitination studies due to the heterogeneous nature of ubiquitinated proteins. Strong cation exchange (SCX) chromatography following tryptic digestion provides excellent separation of ubiquitin-derived peptides, particularly the characteristic Gly-Gly remnant left on modified lysines after trypsin digestion.
Table 2: Comparison of Enrichment Methods for Low-Abundance Proteins
| Method | Principle | Advantages | Limitations |
|---|---|---|---|
| Combinatorial Peptide Ligand Libraries | Affinity enrichment using diverse hexapeptide beads | Reduces dynamic range, concentrates LAP, applicable to various samples | Requires large sample volumes, relatively expensive, single-use |
| Immunoaffinity Enrichment | Antibody-based capture of specific linkages | High specificity, can be linkage-specific, compatible with various detection methods | Limited antibody availability, potential cross-reactivity, co-depletion issues |
| Chromatographic Fractionation | Separation based on physicochemical properties | High binding capacity, various separation modes, scalable | Fraction overlapping, can be time-consuming, sample dilution |
| Glycoprotein Capture | Lectin affinity for glycoproteins | Group-specific, concentrates specific classes, removes non-glycosylated background | Limited to glycosylated targets, non-specific binding |
| Immunosubtraction | Removal of abundant proteins | Reduces signal masking, improves detection limits | Massive co-depletion, sample dilution, restricted to specific sample types |
Cutting-Edge Detection Technologies and Methodologies
Advanced Biosensing Platforms
Emerging biosensing technologies offer unprecedented sensitivity for detecting low-abundance targets without requiring extensive sample preprocessing. Microfluidic-electrochemical biosensors combining AC electrokinetic enrichment with capacitance detection have achieved remarkable detection limits down to 0.1 fM for nucleic acids and fg/mL levels for proteins with turnaround times under one minute [65]. These platforms leverage electrokinetic phenomena to concentrate targets at electrode surfaces, dramatically enhancing signal-to-noise ratios.
CRISPR-based biosensing represents another transformative approach, with representative microfluidic CRISPR sensors demonstrating detection of 20 pfu/mL of purified Ebola RNA within 5 minutes [65]. When adapted for ubiquitin research, these systems could potentially detect transcript-level changes in ubiquitin pathway components or utilize ubiquitin-specific aptamers for direct protein detection. The integration of artificial intelligence with triboelectric nanogenerators (AI-TENGs) further expands the potential for portable, self-powered detection systems suitable for point-of-care applications [65].
Single-Molecule Sequencing for Complex Targets
Single-molecule sequencing technologies have overcome traditional limitations in analyzing repetitive genomic regions, including expanded triplet repeats associated with neurodegenerative diseases. PacBio sequencing has successfully sized C9orf72 repeat expansions ranging into thousands of repeats with superior accuracy compared to Southern blotting, while requiring significantly less input material (3μg vs 20μg) [64]. This approach enables phasing of complex alleles and precise determination of editing outcomes in genetically modified systems.
For ubiquitin research, the ability to sequence through repetitive elements and GC-rich regions makes single-molecule approaches valuable for characterizing ubiquitin locus polymorphisms, editing outcomes in ubiquitin pathway engineering, and transcriptional regulation of ubiquitin genes. The technology's sensitivity to detect impure edited clones even after single-cell sorting (20-80% impurity observed) provides crucial quality control for cell line development [64].
Digital PCR for Quantitative Expression Analysis
Digital PCR (dPCR) provides absolute quantification of nucleic acid targets without requiring standard curves, offering superior precision for low-abundance transcripts. In ubiquitin research, dPCR assays have been developed to differentiate between C9orf72 transcript variants, demonstrating that most neuronal mRNAs (96%) contain exon 1B, while only a small proportion (4%) contain exon 1A [64]. The exceptional sensitivity of dPCR enables detection of rare splice variants and low-level expression changes in ubiquitin pathway components.
The technology's validation using selective exon excision models provides a template for rigorous assay validation in ubiquitin research. dPCR probes spanning specific exon-exon junctions can distinguish between ubiquitin transcript variants with high specificity, enabling researchers to study transcriptional regulation of the ubiquitin system under different physiological and pathological conditions.
Integrated Experimental Protocols for Atypical Ubiquitin Chain Analysis
Comprehensive Workflow for K27-Linked Chain Enrichment and Characterization
The following protocol provides a detailed methodology for studying K27-linked ubiquitination in the context of innate immune signaling:
Step 1: Cell Stimulation and Lysis
- Culture appropriate cell lines (HEK293T or THP-1) and stimulate with viral mimetics (e.g., poly(I:C) at 1μg/mL for 6 hours) or interferon (IFN-α at 1000U/mL for 24 hours) to enhance K27 ubiquitination signaling
- Harvest cells and lyse in NP-40 lysis buffer (50mM Tris-HCl pH 7.5, 150mM NaCl, 1% NP-40) supplemented with fresh 10mM N-ethylmaleimide (NEM) to preserve ubiquitin conjugates
- Clarify lysates by centrifugation at 16,000×g for 15 minutes at 4°C
Step 2: Tandem Enrichment Using CPLL and Immunoprecipitation
- Incubate cleared lysates with CPLL beads (0.5mL beads per 10mg total protein) for 2 hours at 4°C with gentle rotation
- Wash beads extensively with PBS and elute bound proteins using 8M urea in 50mM Tris-HCl (pH 8.0)
- Dilute eluate 1:5 with NP-40 lysis buffer and incubate with K27-linkage specific antibody (2μg per mg protein) overnight at 4°C
- Capture immune complexes with protein A/G beads, wash stringently, and elute with 2× Laemmli buffer for Western analysis or 100mM glycine (pH 2.5) for mass spectrometry
Step 3: Downstream Analysis by Western Blot and Mass Spectrometry
- For Western blot: Separate proteins by SDS-PAGE, transfer to PVDF membranes, and probe with antibodies against targets of interest (MAVS, NEMO, STING)
- For mass spectrometry: Digest eluted proteins with trypsin and analyze by LC-MS/MS using a method optimized for ubiquitin remnant peptide detection (monitoring for GG signature on lysine)
Protocol for Single-Cell Analysis of Ubiquitin Chain Modifications
Recent advances in single-cell proteomics enable investigation of ubiquitin chain heterogeneity at cellular resolution:
Step 1: Single-Cell Sorting and Lysis
- Sort individual cells into 96-well plates containing 5μL lysis buffer (1% SDS, 50mM Tris pH 8.0, 10mM NEM) using a fluorescence-activated cell sorter
- Heat samples at 95°C for 5 minutes to denature proteins and inactivate deubiquitinases
Step 2: Microfluidic Proteomic Sample Preparation
- Process samples using a microfluidic platform for proteomic analysis (commercial systems available)
- Perform reduction, alkylation, and digestion with trypsin/Lys-C mix directly in microfluidic chambers
- Label peptides with tandem mass tags (TMTpro 16-plex) to enable multiplexed analysis
Step 3: Ubiquitin Peptide Enrichment and LC-MS/MS Analysis
- Pool labeled peptides and enrich for ubiquitin remnant peptides using anti-K-ε-GG antibody-conjugated beads
- Fractionate using high-pH reverse-phase chromatography followed by LC-MS/MS on a high-resolution mass spectrometer
- Analyze data using search engines configured to identify ubiquitin signature peptides and quantify changes across conditions
Research Reagent Solutions for Atypical Ubiquitin Studies
Table 3: Essential Research Reagents for Atypical Ubiquitin Chain Studies
| Reagent Category | Specific Examples | Function/Application | Validation Notes |
|---|---|---|---|
| Validated Antibodies | GTX632041, GTX634482, 2575-1-AP, B01-5F2 [64] | Detection of ubiquitin chains by Western blot | Specificity confirmed by knockout controls; not recommended for ICC |
| E3 Ligase Tools | Recombinant TRIM23, RNF185, Parkin | In vitro ubiquitination assays | Specific for K27 (TRIM23, RNF185) and K6 (Parkin) linkages |
| DUB Inhibitors | USP30 inhibitors, OTUD1 probes | Modulating ubiquitin chain stability | Specificity varies; requires empirical testing |
| Mass Spec Standards | Heavy-labeled ubiquitin, K27-linked diubiquitin | Quantification standardization | Essential for absolute quantification studies |
| Cell Line Models | C9orf72 KO iPSCs, Parkin overexpression lines | Pathophysiological context | Provides relevant biological systems for validation |
Visualization of Experimental Workflows and Signaling Pathways
Comprehensive Workflow for Atypical Ubiquitin Chain Analysis
Atypical Ubiquitin Chain Signaling in Innate Immunity
The field of atypical ubiquitin chain research is rapidly evolving from phenomenological observations toward mechanistic understanding, driven by increasingly sophisticated enrichment and detection technologies. The strategies outlined in this technical guide provide a roadmap for investigating the functions of K6, K27, and K29-linked chains despite their challenging low-abundance characteristics. Future advances will likely emerge from several key directions: single-cell ubiquitinomics enabling resolution of cellular heterogeneity; spatial ubiquitin profiling mapping chain distributions within tissues; and dynamic imaging approaches capturing real-time ubiquitination events in living cells. Additionally, the development of highly specific chemical probes for atypical chain types will accelerate both basic research and drug discovery efforts targeting the ubiquitin system.
As these technologies mature, they will undoubtedly reveal new biological functions for atypical ubiquitin chains and expand our understanding of their roles in human health and disease. The integration of sensitive detection methods with sophisticated biological models will continue to decode the complex language of ubiquitin signaling, potentially unlocking new therapeutic opportunities for conditions ranging from cancer to neurodegenerative disorders.
Overcoming Linkage Specificity Issues in E2/E3 Enzyme Assays
The ubiquitin-proteasome system regulates virtually every cellular process, with specificity often dictated by the type of polyubiquitin chain linkage formed during E2/E3-catalyzed ubiquitination. While the functions of K48 and K63 linkages are well-established, research into atypical ubiquitin chains (K6, K27, K29) presents unique experimental challenges due to linkage specificity issues in enzyme assays. These atypical linkages, though less abundant, play critical roles in immune signaling, DNA damage response, and metabolic regulation. This technical guide examines the sources of linkage specificity challenges in E2/E3 enzyme assays and provides detailed methodologies for conducting rigorous research on atypical chain functions, enabling researchers to advance our understanding of these complex post-translational modifications.
The Challenge of Linkage Specificity in Ubiquitin Research
The ubiquitin code's complexity stems from the ability to form diverse polyubiquitin chains through different linkage types. Atypical chains (K6, K27, K29) constitute a minority of cellular ubiquitin linkages but regulate crucial biological pathways:
- K6-linked chains function in DNA damage response and mitophagy, with HUWE1 identified as an E3 capable of modifying substrates with K6-linked chains [66].
- K27-linked chains participate in innate immune regulation and mitophagy, with TRIM family E3s (TRIM23, TRIM27) and RNF185 catalyzing K27 linkages [8].
- K29-linked chains regulate Wnt signaling and mRNA stability, with UFD2 and UFD4 collaborating to synthesize branched K29/K48 chains [24] [12].
Linkage specificity issues arise from several experimental factors:
- E2/E3 promiscuity: Many E2 enzymes and E3 ligases exhibit flexibility in their linkage specificity
- Chain branching: Heterotypic and branched chains complicate linkage-specific analysis
- Technical limitations: Antibody cross-reactivity and mass spectrometry detection challenges
Essential Research Reagents for Atypical Chain Studies
Table 1: Key Research Reagents for Studying Atypical Ubiquitin Linkages
| Reagent Type | Specific Examples | Function in Assays | Considerations |
|---|---|---|---|
| Ubiquitin Mutants | K-to-R (K6R, K27R, K29R) mutants | Identify essential lysines for chain formation | Combinatorial mutants address redundancy [24] |
| E2 Enzymes | UBE2G2 (K48-specific), UBE2S (K11-specific) | Linkage-defined chain initiation and elongation | E2-E3 fusion proteins (gp78RING-Ube2g2) enhance specificity [67] |
| E3 Ligases | HUWE1 (K6), TRIM23 (K27), UFD4 (K29) | Catalyze specific linkage formation | Multiple E3s may collaborate for branched chains [12] |
| Deubiquitinases (DUBs) | Linkage-specific DUBs | Validate chain linkage identity | Limited availability for atypical linkages |
| Detection Tools | Linkage-specific antibodies, Ubi-AQUA-PRM | Identify and quantify specific chains | Variable commercial antibody quality [21] |
Experimental Strategies for Overcoming Specificity Challenges
Genetic Approaches for Pathway Identification
Genetic interaction studies provide powerful tools for identifying pathways regulated by specific ubiquitin linkages:
- Ubiquitin mutant strains: Engineer yeast strains expressing K-to-R ubiquitin mutants at all ubiquitin loci to eliminate specific linkage types while maintaining normal ubiquitin expression levels [24].
- Synthetic Genetic Array (SGA) analysis: Systematically mate ubiquitin mutant strains with gene deletion libraries to identify genetic interactions that reveal functional pathways [24].
- Validation assays: Confirm identified pathways through functional tests (e.g., amino acid import assays for K11 linkages, cell cycle analysis) [24].
Table 2: Optimized Ub-AQUA-PRM Mass Spectrometry Parameters for Atypical Chain Detection
| Parameter | Recommended Setting | Rationale |
|---|---|---|
| Sample Preparation | Rapid urea/thiourea lysis | Preserves native ubiquitin landscape [21] |
| Chromatography | 10-min LC-MS/MS runs | Enables high-throughput screening [21] |
| Quantification | Absolute quantification with heavy labeled peptides | Accurate measurement of low-abundance chains |
| Data Analysis | Targeted extraction of linkage-specific signatures | Focuses sensitivity on atypical linkages |
| Validation | Comparison with linkage-deficient mutants | Confirms specificity of detection |
Ubi-Tagging for Controlled Conjugation
The ubi-tagging approach enables precise control over ubiquitin conjugation for generating defined protein complexes:
Ubi-Tagging Experimental Workflow
Reaction Setup:
- Ubdon construct: Contains free C-terminal glycine with specific lysine mutated to arginine (e.g., K48R) to prevent homodimer formation [67]
- Ubacc construct: Contains corresponding conjugation lysine (e.g., K48) with unreactive C-terminus (ΔGG or His-tag) [67]
- Enzyme system: Specific E1 and E2-E3 fusion proteins (e.g., gp78RING-Ube2g2 for K48 linkages) [67]
Protocol Details:
- Incubate 10μM Fab-Ub(K48R)don with 50μM Rho-Ubacc-ΔGG
- Add 0.25μM E1 and 20μM E2-E3 fusion enzyme
- Reaction time: 30 minutes at room temperature
- Purification: Protein G affinity purification
- Validation: ESI-TOF mass spectrometry confirms complete consumption of starting material and formation of defined product [67]
Design of Experiments (DoE) for Assay Optimization
Traditional one-factor-at-a-time optimization requires >12 weeks, while DoE approaches identify optimal conditions in <3 days [68] [69]:
DoE Implementation Steps:
- Initial screening: Use fractional factorial design to identify significant factors affecting enzyme activity
- Response surface methodology: Determine optimal concentrations and conditions
- Model validation: Confirm predicted optimal conditions experimentally
Key Factors for E2/E3 Assays:
- Buffer composition and pH
- E1, E2, and E3 concentrations
- Ubiquitin and substrate concentrations
- Reaction time and temperature
- ATP and magnesium concentrations
Analytical Methods for Verification
Targeted Proteomics for Chain Linkage Composition
The Ub-AQUA-PRM (Ubiquitin Absolute Quantification by Parallel Reaction Monitoring) method enables comprehensive chain linkage analysis:
Ub-AQUA-PRM Workflow
Protocol Details:
- Tissue processing: Rapid lysis with urea/thiourea buffer to preserve endogenous ubiquitin states [21]
- Peptide preparation: Trypsin digestion generates linkage-specific signature peptides
- LC-MS/MS analysis: 10-minute chromatographic separation enables high-throughput processing
- Absolute quantification: Heavy labeled internal standards for each linkage type
- Data interpretation: Identify tissue-specific enrichment patterns (e.g., K33 enrichment in contractile tissues) [21]
Functional Validation in Physiological Contexts
Case Study: K27 Linkages in Innate Immunity [8]
- Experimental system: Cell-based assays measuring IFN production and NF-κB activation
- E3 identification: TRIM23, TRIM27, RNF185 as K27-specific E3s
- Functional readouts: IRF3 activation, cytokine production, viral replication assays
- Validation approaches: E3 knockdown, linkage-specific ubiquitination assays
Emerging Approaches and Future Directions
Branched Chain Analysis
Branched ubiquitin chains containing atypical linkages present additional complexity:
- Collaborative E3 mechanisms: Pairs of E3s with distinct specificities collaborate to form branched chains (e.g., Ufd4 and Ufd2 for K29/K48 branches) [12]
- Architectural diversity: Branch points can be initiated at distal, proximal, or internal ubiquitins within chains [12]
- Functional specialization: Branched chains can convert non-degradative signals to degradative signals [12]
Single-Chain Analysis Techniques
Advanced methodologies are needed to address:
- Linkage heterogeneity within polyubiquitin chains
- Stoichiometry of branched chains
- Cell-type specific ubiquitin landscapes
Overcoming linkage specificity challenges in E2/E3 enzyme assays requires integrated approaches combining genetic, biochemical, and analytical methods. The methodologies outlined in this guide—including ubi-tagging for controlled conjugation, DoE for assay optimization, and Ub-AQUA-PRM for linkage quantification—provide researchers with robust tools to advance our understanding of atypical ubiquitin chain functions. As these techniques continue to evolve, they will undoubtedly reveal new biological insights and therapeutic opportunities targeting the ubiquitin system.
Distinguishing Heterotypic and Branched Chains from Homotypic Polymers
Ubiquitylation is a crucial post-translational modification that controls diverse cellular processes in eukaryotes, ranging from protein degradation to cell signaling and DNA repair [12]. The versatility of ubiquitin signaling stems from its ability to form diverse polymeric structures through the conjugation of subsequent ubiquitin molecules to one of the seven lysine residues (K6, K11, K27, K29, K33, K48, K63) or the N-terminal methionine (M1) of the preceding ubiquitin moiety [12] [1]. These ubiquitin polymers can be classified into three major categories based on their linkage patterns and architecture. Homotypic chains contain a uniform linkage type throughout the polymer, while heterotypic chains encompass chains with multiple linkage types and can be further divided into mixed chains (each ubiquitin modified at only one site) and branched chains (comprising ubiquitin subunits simultaneously modified on at least two different acceptor sites) [12]. The emerging understanding of heterotypic and branched ubiquitin chains represents a significant expansion of the ubiquitin code, particularly in the context of understudied atypical linkages like K6, K27, and K29, which form the focus of ongoing research efforts.
Structural and Architectural Diversity
The architectural complexity of ubiquitin chains extends far beyond simple linear polymers, creating a sophisticated language for cellular communication.
Defining Chain Topologies
Table 1: Classification of Ubiquitin Chain Architectures
| Chain Type | Structural Definition | Linkage Pattern | Representative Examples |
|---|---|---|---|
| Homotypic | Uniform linkage throughout chain | Single linkage type (e.g., K48-only) | K48-linked chains (proteasomal degradation) [70] |
| Mixed Heterotypic | Multiple linkages, each ubiquitin modified at one site | Sequential ubiquitins with different linkages | M1/K63 mixed chains (NF-κB signaling) [12] |
| Branched Heterotypic | Ubiquitin subunits modified on ≥2 sites simultaneously | Branch points with multiple linkages | K11/K48-branched chains (cell cycle regulation) [71] |
Structural Basis of Branching
Branched ubiquitin chains incorporate at least one ubiquitin molecule that serves as a branch point by being simultaneously modified on two different acceptor sites [12]. This architectural complexity can manifest in several forms: distal branching (initiated at the chain terminus), proximal branching (near the substrate attachment point), or internal branching (within the chain interior) [12]. The specific order of linkage assembly further diversifies chain architecture—for instance, branched K11/K48 chains can be formed by adding K11 linkages to pre-existing K48 chains (as with APC/C) or by adding K48 linkages to pre-formed K11 chains (as with UBR5) [12].
The conformational landscapes of different ubiquitin linkages provide the structural basis for their functional specialization. Single-molecule FRET analyses have revealed that Lys48-linked diUb predominantly adopts compact conformations (~90% high-FRET species), while Lys63- and Met1-linked diUb exist in equilibrium between extended ("open") and more compact ("closed") conformations [72]. These inherent structural dynamics enable selective recognition by ubiquitin-interacting proteins (UbIPs), with ubiquitin-binding domains (UBDs) and deubiquitinases (DUBs) selecting pre-existing conformations rather than inducing structural changes [72].
Synthesis Mechanisms and Enzymatic Assembly
The assembly of branched ubiquitin chains requires precise enzymatic coordination, often involving collaborative efforts between multiple ubiquitination enzymes.
Collaborative E3 Ligase Mechanisms
A predominant mechanism for branched chain synthesis involves collaboration between pairs of E3 ligases with distinct linkage specificities [12]. Several well-characterized examples illustrate this paradigm:
K29/K48 branched chains: In yeast, Ufd4 and Ufd2 collaborate to synthesize branched K29/K48 chains on substrates of the ubiquitin fusion degradation (UFD) pathway. Ufd4 initially assembles K29-linked chains, which are then recognized by Ufd2 through two specific loops in its N-terminal domain, enabling the addition of K48 linkages to create branch points [12].
K48/K63 branched chains: During NF-κB signaling, TRAF6 and HUWE1 cooperate to produce branched K48/K63 chains. TRAF6 synthesizes K63-linked chains that are subsequently recognized by HUWE1 through its UIM and UBA domains, leading to the attachment of K48 linkages [12]. Similarly, in apoptotic regulation, ITCH and UBR5 collaborate—ITCH first modifies the pro-apoptotic regulator TXNIP with K63-linked chains, which are then bound by the UBA domain of UBR5, enabling UBR5 to attach K48 linkages and create proteasome-targeting branched K48/K63 chains [12].
Single-E3 Mechanisms
Some individual E3 ligases possess intrinsic capacity to synthesize branched chains, either by recruiting multiple E2 enzymes with distinct specificities or through inherent catalytic flexibility:
APC/C-mediated branching: The anaphase-promoting complex/cyclosome (APC/C), a multisubunit RING E3, cooperates with UBE2C and UBE2S to form branched K11/K48 chains on mitotic substrates. UBE2C initially attaches short chains containing mixed K11, K48, and K63 linkages, followed by UBE2S extending these primers with multiple K11 linkages to create branched architectures [12] [70].
HECT E3 versatility: Several HECT-family E3s, including WWP1, UBE3C, and the bacterial NleL, can assemble branched chains (K48/K63, K29/K48, and K6/K48, respectively) with a single E2 enzyme [12]. These E3s often contain non-covalent ubiquitin-binding sites within or adjacent to their catalytic HECT domains that may facilitate branch point recognition and utilization [12].
Figure 1: Enzymatic Pathways for Branched Ubiquitin Chain Assembly. Branched chains can be synthesized through collaborative mechanisms involving multiple E3 ligases or via single E3s that recruit multiple E2 enzymes with distinct linkage specificities.
Functional Specialization and Biological Consequences
Branched ubiquitin chains expand the functional repertoire of ubiquitin signaling by enabling sophisticated regulatory mechanisms beyond the capabilities of homotypic chains.
Enhanced Proteasomal Targeting
A key functional advantage of branched ubiquitin chains lies in their enhanced efficacy in proteasomal targeting. Research has demonstrated that while homotypic K11-linked chains bind weakly to the proteasome and fail to stimulate degradation, heterotypic K11/K48-branched chains exhibit strong proteasome binding and efficiently target substrates like cyclin B1 for degradation [70]. This distinction arises from fundamental differences in how the proteasome recognizes different chain topologies—the proteasome and its shuttling factors preferentially bind K48-linked chains over K11 linkages, but efficiently engage heterotypic K11/K48-branched architectures [70].
Recent technological advances using the UbiREAD platform have further elucidated the degradation code of branched ubiquitin chains, revealing that branched chains are not simply the sum of their parts but exhibit functional hierarchy where substrate-anchored chain identity determines degradation behavior [73]. K48 chains with three or more ubiquitins trigger rapid degradation within minutes, while K63-ubiquitinated substrates are rapidly deubiquitinated rather than degraded [73].
Signaling Regulation and Protein Quality Control
Branched ubiquitin chains serve critical functions in cell cycle regulation and protein quality control. During mitosis, K11/K48-branched chains assembled by the APC/C ensure the precise temporal degradation of cell cycle regulators [71]. Additionally, endogenous K11/K48-branched chains target misfolded nascent polypeptides and pathological Huntingtin variants for rapid proteasomal clearance, establishing their essential role in maintaining proteostasis and preventing protein aggregation [71].
The functional specialization of branched chains extends to immune signaling pathways, where they contribute to both activation and attenuation of inflammatory responses. For instance, the conversion of non-degradative K63-linked signals to degradative K48/K63-branched chains provides an efficient mechanism for regulating the activation and inactivation of signaling proteins in pathways like NF-κB signaling [12] [33].
Methodologies for Analysis and Detection
Advancing our understanding of branched ubiquitin chains requires sophisticated methodological approaches capable of distinguishing complex chain architectures.
Linkage-Specific Reagents and Antibodies
The development of linkage-specific reagents has been instrumental in deciphering branched ubiquitin chain functions. A breakthrough innovation came with the engineering of a K11/K48-bispecific antibody using knobs-into-holes heterodimerization technology [71]. This antibody functions as a coincidence detector that gains avidity from simultaneously recognizing both K11- and K48-linkages, enabling specific detection of K11/K48-branched conjugates while showing minimal affinity for homotypic K11- or K48-linked chains or other branched variants (K11/K63, K48/K63, or M1/K63) [71].
Table 2: Key Research Reagents for Branched Ubiquitin Chain Analysis
| Reagent / Method | Specificity / Application | Key Features | References |
|---|---|---|---|
| K11/K48-bispecific antibody | K11/K48-branched chains | Functions as coincidence detector; recognizes endogenous conjugates | [71] |
| Linkage-specific DUBs | Specific linkage cleavage | AMSH (K63-specific); OTUB1 (K48-specific) | [70] [72] |
| UbiREAD platform | Degradation kinetics | Monitors cellular degradation and deubiquitination at high temporal resolution | [73] |
| Tandem UBD domains | Ubiquitinated protein enrichment | Higher affinity enrichment of ubiquitinated proteins from native sources | [26] |
| Single-molecule FRET | Conformational dynamics | Resolves distinct conformational states of different diUb linkages | [72] |
Mass Spectrometry and Proteomic Approaches
Mass spectrometry-based methods have become cornerstone techniques for ubiquitin research. Advanced approaches now enable the characterization of ubiquitination sites, linkage types, and chain architecture through di-glycine remnant identification after trypsin cleavage [26] [1]. The development of Ub tagging systems, such as His-tagged or Strep-tagged ubiquitin, allows affinity purification of ubiquitinated proteins, though these methods may introduce artifacts and cannot be applied to native tissues [26]. For endogenous ubiquitination studies, antibodies with general ubiquitin specificity (e.g., P4D1, FK1/FK2) or linkage-selective antibodies enable enrichment of ubiquitinated proteins from native sources, including clinical samples [26].
Conformational and Biophysical Analysis
Single-molecule FRET has provided crucial insights into the conformational dynamics of different ubiquitin linkages. This approach has revealed that K48-linked diUb exists predominantly in compact conformations, while K63- and M1-linked diUb adopt more extended conformations [72]. These inherent structural dynamics directly influence recognition by UbIPs—DUBs like AMSH-LP and USP21 select pre-existing open conformations, while UBDs like the NEMO UBAN domain bind compact conformations [72]. Such biophysical analyses help explain the molecular basis for specificity in recognizing different chain topologies.
Figure 2: Experimental Workflow for Branched Ubiquitin Chain Analysis. Comprehensive analysis of branched chains requires specialized approaches for sample preparation, enrichment of ubiquitinated proteins, analytical readouts, and functional interpretation.
Research Challenges and Future Perspectives
Despite significant advances, several challenges remain in the comprehensive analysis of branched ubiquitin chains. The low stoichiometry of ubiquitination under physiological conditions complicates detection, while the complexity of possible chain architectures (varying in length, linkage combinations, and branch point locations) creates an enormous analytical space [26]. Future methodological developments need to address the limited availability of specific reagents for atypical linkages like K6, K27, and K29, particularly bispecific reagents capable of detecting branched chains incorporating these linkages [33].
The development of more sophisticated proteomic approaches, improved mass spectrometry capabilities for detecting branched peptides, and advanced structural biology techniques will be essential for mapping the complete landscape of branched ubiquitin signals. Furthermore, understanding the spatial and temporal regulation of branched chain assembly and disassembly in cellular contexts represents a critical frontier, particularly for elucidating the roles of atypical ubiquitin linkages in disease pathogenesis and their potential as therapeutic targets.
As research methodologies continue to evolve, the integration of biochemical, proteomic, and biophysical approaches will be essential for deciphering the complex language of branched ubiquitin signaling and its implications for cellular regulation and disease mechanisms.
Among the eight homotypic ubiquitin chain linkages, lysine 27 (K27)-linked polyubiquitin stands out for its unique resistance to deubiquitinases (DUBs) [14] [74]. This characteristic distinguishes it from both the well-characterized proteolytic K48-linked chains and the non-proteolytic K63-linked chains, placing it within the less-understood category of "atypical" ubiquitin linkages that also includes K6, K11, K29, and K33 [33]. The DUB resistance of K27 linkages presents a significant biological problem: how can this important ubiquitin signal be regulated if it cannot be easily removed by conventional erasers of the ubiquitin code? Solving this problem is critical not only for understanding fundamental cell biology but also for developing therapeutic strategies for related diseases.
K27-linked ubiquitination represents less than 1% of total ubiquitin conjugates in human cells [74], which may be partly explained by structural studies showing that K27 is the least solvent-exposed lysine residue in ubiquitin [74]. Despite its low abundance, recent research has revealed that K27-linked ubiquitylation is essential for proliferation of human cells [74] and plays important roles in diverse cellular processes including mitochondrial quality control, DNA damage repair, innate immune signaling, and cell cycle progression [14] [74] [33]. This review examines the molecular basis of K27 linkage resistance to DUBs, explores its cellular functions, and discusses experimental approaches to study this enigmatic ubiquitin signal.
Structural and Biochemical Basis of K27 Linkage DUB Resistance
Unique Structural Properties of K27-Linked Di-Ubiquitin
The DUB resistance of K27 linkages is rooted in the unique structural properties of K27-linked polyubiquitin chains. Comprehensive structural characterization using NMR spectroscopy, small-angle neutron scattering (SANS), and in silico ensemble modeling has revealed that K27-linked di-ubiquitin (K27-Ub2) adopts a compact conformation with restricted accessibility [14].
Unlike other ubiquitin linkages where significant noncovalent interdomain contacts occur between ubiquitin units, K27-Ub2 exhibits minimal noncovalent interactions between the distal and proximal ubiquitin moieties [14]. NMR chemical shift perturbation (CSP) analyses demonstrate that the distal Ub in K27-Ub2 shows the smallest CSPs of all Ub2s studied, indicating a lack of stable interdomain contacts [14]. In contrast, the proximal Ub of K27-Ub2 displays the largest and most widespread CSPs among all Ub2s [14], suggesting that the isopeptide linkage itself imposes structural constraints that limit DUB accessibility.
The K27 linkage site is particularly buried within the ubiquitin structure. Comparative analysis of all lysine linkage sites reveals that K27 is the least solvent-exposed lysine residue in ubiquitin [74], which may explain why most DUBs display poor activity toward K27 linkages. This structural inaccessibility creates a physical barrier that prevents the catalytic domains of many DUBs from properly engaging with and cleaving the isopeptide bond.
DUB Profiling Reveals Broad Resistance Patterns
Experimental screening of K27-Ub2 against a panel of DUBs representing different families has demonstrated its exceptional resistance to cleavage. As shown in Table 1, K27-Ub2 resists disassembly by multiple DUB families, including linkage-nonspecific enzymes that efficiently cleave other ubiquitin linkages [14].
Table 1: DUB Resistance Profile of K27-Ub2 Compared to Other Linkages
| DUB Enzyme | DUB Family | Preferred Linkage | K27-Ub2 Cleavage | K48-Ub2 Cleavage | K63-Ub2 Cleavage |
|---|---|---|---|---|---|
| Cezanne | OTU | K11-specific | Resistant | Not preferred | Not preferred |
| OTUB1 | OTU | K48-specific | Resistant | Efficient | Not preferred |
| AMSH | JAMM | K63-specific | Resistant | Not preferred | Efficient |
| USP2 | USP | Linkage-nonspecific | Resistant | Efficient | Efficient |
| USP5 (IsoT) | USP | Linkage-nonspecific | Resistant | Efficient | Efficient |
| Ubp6 | USP | Linkage-nonspecific | Resistant | Efficient | Efficient |
Notably, K27 was the only linkage that resisted cleavage by USP5 (IsoT), a DUB known for its ability to cleave all types of ubiquitin chains [14]. K27-Ub2 also resisted disassembly by the proteasome-associated DUB Rpn11 in reconstituted proteasome lid core complex [14]. This broad DUB resistance means that K27-Ub2 can function as a competitive inhibitor of DUB activity toward other linkages [14], suggesting potential regulatory crosstalk between different ubiquitin signaling pathways.
Cellular Functions of K27-Linked Ubiquitination
Essential Roles in Cell Viability and Nuclear Processes
Recent studies using conditional ubiquitin replacement strategies have demonstrated that K27-linked ubiquitylation is essential for human cell proliferation [74]. When endogenous ubiquitin was replaced with a Ub(K27R) mutant that cannot form K27 linkages, cells showed severely impaired colony formation ability—comparable to the defect imposed by ablation of the essential K48 linkage [74].
K27-linked ubiquitination is predominantly a nuclear modification whose ablation deregulates nuclear ubiquitylation dynamics and impairs cell cycle progression [74]. This function is epistatic with inactivation of the ATPase p97/VCP, a key regulator of protein homeostasis [74]. The p97-proteasome pathway model substrate Ub(G76V)-GFP is directly modified by K27-linked ubiquitylation, and disabling the formation of K27-linked ubiquitin signals or blocking their recognition impedes substrate turnover at the level of p97 function [74].
Regulation of Immune Signaling Pathways
K27-linked ubiquitination plays important regulatory roles in innate and adaptive immunity. In antiviral innate immune signaling, the E3 ligase TRIM23 conjugates K27-linked chains to NEMO (NF-κB essential modulator), and this modification is required for induction of NF-κB and IRF3 upon RIG-I-like receptor (RLR) signaling activation [33]. K27-linked chains on NEMO subsequently serve as an interaction platform for other regulatory factors [33].
In adaptive immunity, a novel K27-linked ubiquitination mechanism regulates Th17 cell differentiation and autoimmunity [75]. The HECT E3 ubiquitin ligase Nedd4 binds to the PPLY motif within the ligand binding domain of RORγt (the master transcription factor controlling Th17 cell differentiation) and targets RORγt at K112 for K27-linked polyubiquitination, thereby enhancing its activity [75]. Disruption of this pathway impairs pathogenic and non-pathogenic Th17 responses and ameliorates experimental autoimmune encephalomyelitis (EAE), a mouse model of multiple sclerosis [75].
Mitochondrial Quality Control
K27-linked ubiquitination serves as an important marker in mitochondrial quality control. Mitochondrial trafficking protein Miro1 is modified with K27-linked chains, which slows down its degradation by the proteasome and acts as a marker of mitochondrial damage [14]. This positioning of K27 linkages at the intersection of mitochondrial homeostasis and protein turnover regulation highlights the diverse functional roles of this ubiquitin linkage.
Experimental Approaches to Study K27 Linkages
Methodologies for Producing K27-Linked Ubiquitin Chains
Studying K27 linkages has been challenging due to the lack of linkage-specific ubiquitin-conjugating enzymes. Several innovative approaches have been developed to overcome this limitation:
Non-enzymatic chemical assembly: A strategy utilizing mutually orthogonal removable amine-protecting groups Alloc and Boc has been developed to assemble fully natural K27-Ub2 with native isopeptide linkages free of any mutations [14]. This chemical biology approach enables production of structurally authentic K27-linked chains for biochemical and structural studies.
DUB-resistant linear ubiquitin chains: Expression of linear ubiquitin chains lacking internal "GG" motifs (necessary for isopeptide bond cleavage by DUBs) in Drosophila melanogaster provides a tool to study DUB-resistant polyubiquitin species [76]. These linear chains are rapidly modified with different linkages in vivo, including K27, K48, and K63 linkages, forming various branched chain topologies [76].
Linkage-specific antibodies: Development of K27 linkage-specific antibodies enables enrichment and detection of proteins modified with K27-linked ubiquitin chains [26] [75]. These reagents facilitate the study of endogenous K27 ubiquitination under physiological conditions.
Table 2: Key Research Reagents for Studying K27-Linked Ubiquitination
| Reagent/Tool | Type | Key Features/Applications | Considerations |
|---|---|---|---|
| K27-Ub2 (non-enzymatic assembly) | Chemically synthesized di-ubiquitin | Native isopeptide linkage; For structural studies (NMR, SANS) and biochemical DUB assays | No mutations; Structurally authentic; Limited chain length |
| Ub(K27R) mutant | Ubiquitin point mutant | Abrogates K27-linked ubiquitylation; Used in cellular replacement studies | May affect other linkage formation; Potential compensatory mechanisms |
| K27 linkage-specific antibody | Immunological reagent | Enriches endogenously ubiquitinated proteins with K27 linkages; IHC, WB applications | Specificity validation required; Potential cross-reactivity |
| DUB-resistant linear Ub6 chains | Recombinant polyubiquitin | Resists cleavage by DUBs; Studies of unanchored polyubiquitin fate in vivo | Linear backbone with mixed/branched linkages; Not homotypic K27 chains |
| UCHL3 | K27-linkage specific binder | Recognizes and binds K27 linkages; Functional interference studies | May block K27 signal decoding; Overexpression effects |
Analytical Techniques for Characterization
Multiple biophysical and biochemical techniques have been employed to characterize K27-linked ubiquitin chains:
NMR spectroscopy: Provides atom-specific information about structural dynamics and interdomain interactions in K27-Ub2 [14]. Chemical shift perturbation analysis reveals unique features of both distal and proximal ubiquitin units [14].
Small-angle neutron scattering (SANS): Offers solution-based structural information about the overall architecture and conformational ensemble of K27-Ub2 [14].
In silico ensemble modeling: Computational approaches complement experimental structural data to generate dynamic models of K27-Ub2 conformations [14].
Mass spectrometry-based proteomics: Advanced MS methods enable identification of ubiquitination sites and linkage type characterization [26]. Ubiquitin tagging-based approaches (e.g., His-tagged Ub) and ubiquitin antibody-based approaches allow enrichment and profiling of ubiquitinated substrates [26].
Figure 1: Experimental Workflow for K27 Linkage Research. This diagram outlines the integrated approaches for producing, detecting, and functionally characterizing K27-linked ubiquitin chains, highlighting the multidisciplinary nature of ubiquitin research.
DUB Resistance Mechanisms and Biological Implications
Molecular Basis of Cleavage Resistance
The resistance of K27 linkages to DUB cleavage stems from multiple structural factors:
Steric hindrance: The compact conformation of K27-Ub2 creates physical barriers that prevent optimal positioning of the isopeptide bond within the catalytic cleft of most DUBs [14].
Sequence recognition limitations: Many DUBs have evolved to recognize specific structural features around linkage sites that are absent or altered in K27-linked chains.
Solvent inaccessibility: The buried nature of the K27 linkage site within the ubiquitin structure limits enzymatic access [74].
This resistance profile is not absolute, however, as some DUBs can cleave K27 linkages, suggesting specialized adaptations in these enzymes. Identifying the specific DUBs that efficiently cleave K27 linkages remains an active area of research.
Cellular Fate of DUB-Resistant Chains
The persistence of K27-linked ubiquitin chains in cells necessitates alternative clearance mechanisms beyond DUB-mediated disassembly. Research in Drosophila models has revealed that unanchored polyubiquitin that cannot be cleaved by DUBs is degraded by the proteasome, at least in part through the assistance of VCP (p97) and its cofactor p47 [76]. Additionally, these DUB-resistant unanchored polyubiquitin chains can be conjugated en bloc to other proteins in vivo [76], suggesting an alternative pathway for their metabolism and function.
These findings indicate that unanchored polyubiquitin species need not be intrinsically toxic; they can be controlled independently of DUB-based disassembly through degradation or conjugation onto other proteins [76]. This challenges the prevailing view that accumulation of unanchored chains is necessarily detrimental to cellular homeostasis.
Therapeutic Implications and Future Perspectives
The DUB resistance of K27 linkages presents both challenges and opportunities for therapeutic development. On one hand, the difficulty in naturally reversing this signal suggests that K27-linked ubiquitination may represent a particularly stable regulatory modification that could be exploited for long-term modulation of cellular processes. On the other hand, the identification of specific DUBs capable of cleaving K27 linkages might reveal new therapeutic targets for diseases involving dysregulated K27 ubiquitination.
Recent research has demonstrated that targeting Nedd4, the E3 ligase responsible for K27-linked ubiquitination of RORγt in Th17 cells, attenuates pathogenic immune responses in multiple sclerosis models [75]. This proof-of-concept suggests that modulation of K27-linked ubiquitination pathways represents a promising therapeutic strategy for autoimmune diseases.
Future research directions should focus on:
- Identifying specific DUBs that cleave K27 linkages and their regulatory mechanisms
- Developing more specific tools for manipulating K27 linkages in cellular and animal models
- Elucidating the role of branched ubiquitin chains containing K27 linkages
- Exploring cross-talk between K27 ubiquitination and other post-translational modifications
- Investigating the potential of K27 linkage pathways as therapeutic targets in cancer, neurodegenerative diseases, and autoimmune disorders
Figure 2: K27 Ubiquitination in Cellular Function and Disease. This diagram illustrates the diverse cellular processes, molecular interactions, and pathological associations of K27-linked ubiquitination, highlighting its multifaceted roles in health and disease.
The DUB resistance of K27-linked ubiquitin chains represents both a fascinating biological problem and an opportunity to understand the complexity of ubiquitin signaling. The unique structural features of K27 linkages that confer resistance to most DUBs have shaped the evolution of specialized cellular functions for this ubiquitin signal, particularly in processes requiring stable regulatory modifications such as cell cycle control, immune regulation, and mitochondrial quality control.
While significant progress has been made in understanding the structural basis of K27 linkage DUB resistance and its cellular functions, many questions remain. The development of innovative chemical and genetic tools has enabled researchers to begin deciphering the K27 ubiquitin code, but complete understanding will require integrated approaches combining structural biology, proteomics, and cell biology. As research methodologies continue to advance, the coming years will likely yield new insights into how cells leverage the unique properties of K27 linkages for specialized regulatory functions and how these pathways might be targeted for therapeutic benefit.
The case of K27 linkages exemplifies the broader principle that the diversity of ubiquitin chain architectures enables the ubiquitin system to regulate an extraordinary range of cellular processes with exquisite specificity. Solving the DUB resistance problem of K27 linkages not only advances our understanding of this specific pathway but also provides conceptual frameworks for investigating other atypical ubiquitin modifications in health and disease.
In the rapidly evolving field of ubiquitin research, the study of atypical ubiquitin chain linkages—K6, K27, and K29—presents unique challenges for specificity validation. These less-abundant chain types regulate crucial cellular processes from mitophagy to immune signaling, yet their study is often hampered by reagent cross-reactivity and a historical lack of robust detection tools. Establishing rigorous validation protocols for antibodies, probes, and affinity reagents is therefore paramount to advancing our understanding of these atypical chains' biological functions. This technical guide provides a comprehensive framework for validating reagent specificity within the context of K6, K27, and K29 ubiquitin chain research, incorporating current structural insights and methodological approaches to ensure data reliability and reproducibility.
The Specificity Challenge in Atypical Ubiquitin Research
The ubiquitin code's complexity stems from the ability of ubiquitin to form polymers through eight different linkage types, all utilizing the same 76-amino acid protein. This structural similarity creates significant challenges for achieving linkage specificity. For the atypical linkages K6, K27, and K29, these challenges are particularly pronounced due to their lower cellular abundance and the relative novelty of research tools.
K6-linked chains have been implicated in mitophagy and DNA damage response, with recent structural studies revealing that the NZF domain of TAB2 unexpectedly recognizes both K6 and K63 linkages with similar binding mechanisms [77]. K27 linkages exhibit unique biochemical properties, including remarkable resistance to deubiquitinases (DUBs)—a characteristic that distinguishes them from all other chain types [14]. K29-linked chains participate in Wnt signaling pathways and have been found to exist within mixed or branched chains containing other linkages [10] [12]. This inherent propensity for forming heterotypic chains further complicates specificity validation.
The emergence of non-antibody affinity reagents such as Affimers has provided new opportunities for specific recognition, with crystal structures revealing how these scaffolds achieve linkage specificity through dimerization to create two ubiquitin-binding surfaces with defined spatial orientations [11]. Nevertheless, rigorous validation remains essential, as even highly specific reagents can exhibit cross-reactivity under certain conditions.
Core Principles of Specificity Validation
Multi-Assay Correlation
Relying on a single validation method is insufficient for establishing reagent specificity. Instead, researchers should employ a complementary suite of techniques that evaluate binding characteristics under different conditions. This multi-assay approach provides orthogonal validation, where consistent results across different platforms significantly strengthen specificity claims.
For atypical ubiquitin chain reagents, the validation pipeline should include both quantitative binding assays (ITC, SPR) and application-specific functional tests (western blotting, immunofluorescence). Discrepancies between these assays can reveal important limitations, as demonstrated by the K33-specific Affimer that showed binding by isothermal titration calorimetry (ITC) at high concentrations but failed to detect signals in western blotting at lower concentrations [11].
Structural Understanding
Interpreting validation data requires fundamental knowledge of ubiquitin chain structural biology. The conformational flexibility of different linkages significantly impacts reagent binding. For example, K27-Ub2 exhibits minimal non-covalent interdomain contacts and the largest chemical shift perturbations in NMR studies, reflecting unique dynamic properties that influence recognition [14]. In contrast, K29-linked diubiquitin adopts an extended conformation that exposes hydrophobic patches on both ubiquitin moieties [10].
Understanding these structural characteristics helps explain and predict cross-reactivity patterns. The structural basis for dual K6/K63 specificity in TAB2-NZF, for instance, lies in the flexible C-terminal region of the distal ubiquitin, which accommodates both linkage types with similar binding modes [77].
Experimental Validation Platforms: Methods and Protocols
Quantitative Binding Assays
Isothermal Titration Calorimetry (ITC) ITC provides comprehensive thermodynamic profiling of ubiquitin chain interactions by directly measuring heat changes during binding events. This label-free method in solution avoids immobilization artifacts and yields stoichiometry (n), binding affinity (KD), and thermodynamic parameters (ΔH, ΔS).
Protocol for Affimer-diUb ITC [11]:
- Prepare Affimer and diUb solutions in identical buffer (e.g., 20 mM HEPES, 150 mM NaCl, pH 7.5)
- Degas both solutions to eliminate air bubbles
- Load 300 μL of 50-100 μM Affimer into the sample cell
- Fill syringe with 1-2 mM diUb solution
- Program instrument: 25-30 injections of 1-2 μL each with 120-180s intervals
- Run control experiment by injecting diUb into buffer alone
- Analyze data using independent binding model; note that Affimers may show 2:1 (affimer:diUb) stoichiometry indicating dimerization
Surface Plasmon Resonance (SPR) SPR offers superior sensitivity for kinetic analysis, enabling determination of association (kon) and dissociation (koff) rates that critically influence specificity.
Protocol for TAB2-NZF SPR [77]:
- Immobilize ubiquitin chains on CMS sensor chip via amine coupling to ~100-500 response units
- Establish concentration series of analyte (TAB2-NZF) in HBS-EP buffer (0.01 M HEPES, 0.15 M NaCl, 3 mM EDTA, 0.005% v/v Surfactant P20, pH 7.4)
- Program kinetic titration: contact time 60-120s, dissociation time 120-300s, flow rate 30 μL/min
- Regenerate surface with 10-50 mM NaOH or glycine pH 2.0-3.0
- Analyze sensograms using 1:1 Langmuir binding model or more complex models as needed
Table 1: Representative Binding Data for Atypical Ubiquitin Chain Reagents
| Reagent | Target | Assay | KD | Stoichiometry | Cross-reactivities |
|---|---|---|---|---|---|
| TAB2-NZF [77] | K6-Ub2 | SPR | Not specified | 1:1 | K63-Ub2 |
| TAB2-NZF [77] | K63-Ub2 | SPR | Not specified | 1:1 | K6-Ub2 |
| K6 Affimer [11] | K6-Ub2 | ITC | High affinity | 0.46 (2:1 complex) | Minimal, weak with tetraUb |
| K33 Affimer [11] | K33-Ub2 | ITC | High affinity | 0.44 (2:1 complex) | K11-linked chains |
Application-Specific Validation
Western Blotting Linkage-specific western blotting represents a fundamental application test that evaluates specificity in a denaturing but immobilized format.
Protocol for Affimer Western Blotting [11]:
- Separate 50-100 ng of purified diUb chains of all linkage types by SDS-PAGE (4-12% Bis-Tris)
- Transfer to PVDF membrane using standard protocols
- Block with 5% non-fat milk in TBST for 1 hour
- Incubate with site-specifically biotinylated Affimer (1-2 μg/mL) in blocking buffer overnight at 4°C
- Wash 3× with TBST, 5 minutes each
- Incubate with streptavidin-HRP (1:5000) for 1 hour
- Develop with ECL reagent and image
- Expected result: Strong signal only for cognate linkage with minimal off-target detection
Deubiquitinase (DUB) Sensitivity Assays DUB resistance profiling provides functional validation of linkage identity, particularly valuable for K27 chains that show unique resistance profiles.
Protocol for DUB Sensitivity Screening [14]:
- Prepare 5-10 μg of each diUb linkage in DUB assay buffer (50 mM Tris, 150 mM NaCl, 1 mM DTT, pH 7.5)
- Incubate with 0.5-1 μM of various DUBs (USP2, USP5, Ubp6, OTUB1, AMSH, Cezanne) for 30-60 minutes at 37°C
- Stop reaction with SDS-PAGE loading buffer
- Analyze cleavage by western blotting or Coomassie staining
- Expected result: K27-Ub2 resists cleavage by most DUBs including linkage-nonspecific USPs
Table 2: DUB Sensitivity Profile Across Ubiquitin Linkages
| Linkage | USP2 | USP5 | Ubp6 | OTUB1 | AMSH | Cezanne |
|---|---|---|---|---|---|---|
| K6 | Sensitive | Sensitive | Sensitive | Sensitive | Sensitive | Not specified |
| K27 | Resistant | Resistant | Resistant | Resistant | Resistant | Resistant |
| K29 | Partially resistant | Sensitive | Partially resistant | Sensitive | Sensitive | Not specified |
| K48 | Sensitive | Sensitive | Sensitive | Sensitive | Resistant | Resistant |
| K63 | Sensitive | Sensitive | Sensitive | Resistant | Sensitive | Resistant |
Structural Validation Techniques
X-ray Crystallography High-resolution crystal structures provide the ultimate validation of binding mechanisms and reveal the structural basis for linkage specificity.
Protocol for Crystallizing Affimer-diUb Complexes [11]:
- Purify Affimer and diUb to homogeneity using size exclusion chromatography
- Form complex by incubating at 1:1.2 molar ratio (Affimer:diUb) on ice for 30 minutes
- Screen crystallization conditions using commercial screens (e.g., Hampton Research)
- Optimize hits: K6 affimer-K6-Ub2 complex crystallized in 0.1 M sodium acetate pH 4.5, 28% PEG 4000
- Flash-cool crystals in liquid nitrogen with appropriate cryoprotectant
- Collect data at synchrotron source, process with HKL-3000 or similar
- Solve structure by molecular replacement using known Ub and Affimer structures
Nuclear Magnetic Resonance (NMR) Spectroscopy Solution-state NMR captures the dynamic properties of ubiquitin chains that influence reagent recognition.
Protocol for NMR Analysis of Ubiquitin Chains [14]:
- Prepare 15N-labeled ubiquitin chains in NMR buffer (20 mM sodium phosphate, 50 mM NaCl, 1 mM DTT, pH 6.5)
- Collect 1H-15N HSQC spectra at 25-30°C
- Assign chemical shifts using standard ubiquitin assignments as starting point
- Calculate chemical shift perturbations (CSPs) using formula: CSP = √((ΔδHN)² + (ΔδN/5)²)
- Map CSPs to ubiquitin surface to identify interaction interfaces
- Note: K27-Ub2 shows largest CSPs in proximal ubiquitin with minimal distal perturbations
Special Considerations for Atypical Linkages
K6-Linked Chains
The discovery that TAB2-NZF binds both K6 and K63 linkages necessitates extra validation steps for K6-specific reagents [77]. Always include K63 chains in cross-reactivity panels, and employ structural insights to guide reagent design. The similar binding mechanisms for K6 and K63 result from C-terminal flexibility in the distal ubiquitin, suggesting that reagents targeting more constrained epitopes might achieve better specificity.
K27-Linked Chains
K27 linkages present both challenges and opportunities for validation. Their unique DUB resistance profile provides a functional validation signature unmatched by other linkages [14]. Capitalize on this by including DUB sensitivity as a standard validation step. Additionally, K27 chains exhibit distinct NMR signatures with widespread chemical shift perturbations in the proximal ubiquitin, offering a biophysical validation method.
K29-Linked Chains
The tendency of K29 linkages to exist within mixed or branched chains complicates validation [10] [12]. Always test reagents against heterotypic chains containing K29 linkages, and be cautious when interpreting cellular data, as signals may originate from branched rather than homotypic chains. The extended conformation of K29-diUb exposes hydrophobic patches on both ubiquitin moieties, which influences binding domain recognition [10].
Visualization of Validation Workflows and Signaling Context
Validation Workflow for Ubiquitin Reagents
Atypical Ubiquitin Chains in Immune Signaling
Research Reagent Solutions
Table 3: Essential Research Reagents for Atypical Ubiquitin Research
| Reagent Type | Specific Examples | Function/Application | Source/Reference |
|---|---|---|---|
| Linkage-Specific diUbiquitin | K6, K27, K29 diUb | Positive controls for specificity validation; DUB substrate assays | LifeSensors [78] |
| Activity-Based Probes | K6/K27/K29 diUb-VME | DUB activity profiling; linkage specificity screening | UbiQ [79] |
| Affimer Reagents | K6-specific Affimer | Alternative to antibodies; western blot, pull-downs, microscopy | Michel et al. [11] |
| Ubiquitin Binding Domains | TAB2-NZF, TRABID-NZF | Natural linkage-specific interactors; comparative specificity studies | PMC [77] |
| E3 Ligase Tools | HUWE1, RNF144A/B, TRIM23 | Chain assembly enzymes; cellular model validation | Michel et al. [11] |
Validating the specificity of reagents for atypical ubiquitin chains demands an integrated, multi-platform approach that combines quantitative biophysics with functional assays and structural insights. The unique biochemical properties of K6, K27, and K29 linkages—from K6/K63 cross-reactivity to K27 DUB resistance—necessitate specialized validation strategies beyond standard antibody testing. By implementing the comprehensive framework outlined in this guide, researchers can advance our understanding of these enigmatic ubiquitin signals with greater confidence and reliability, ultimately illuminating their roles in cellular regulation and disease pathogenesis.
Technical Pitfalls in Differentiating Atypical Chains from Common Modifications
The study of atypical ubiquitin chains (K6-, K27-, and K29-linkages) presents unique technical challenges that distinguish them from the well-characterized K48 and K63-linked chains. These low-abundance modifications play critical roles in chromosome biology, epigenome integrity, and stress response pathways, yet their accurate identification and functional characterization are hampered by methodological limitations. This technical guide examines the core pitfalls in differentiating these atypical chains from common modifications and provides detailed experimental frameworks to address these challenges. Through systematic analysis of current methodologies, quantitative profiling, and visualization of complex ubiquitination pathways, we offer researchers a comprehensive toolkit for advancing the study of these enigmatic post-translational modifications within the broader context of ubiquitin chain function research.
Ubiquitination represents one of the most versatile post-translational modifications, regulating virtually every cellular process through a complex code of monomeric and polymeric ubiquitin signals. While K48- and K63-linked chains have been extensively characterized for their roles in proteasomal degradation and signaling transduction respectively, the so-called "atypical" chains (K6, K27, and K29-linked) have remained enigmatic due to technical challenges in their specific detection and manipulation [80]. These chains typically constitute less than 0.5% of total cellular ubiquitin populations, creating substantial detection and characterization hurdles [62]. Recent advances have begun to illuminate their specialized functions: K29-linked ubiquitylation is strongly associated with chromosome biology and essential for proteasomal degradation of the H3K9 methyltransferase SUV39H1 [81] [62]; K27-linkages are critical for cell proliferation and associated with p97 activity in the nucleus [62]; while K6-linked chains mobilize p97/VCP and the proteasome to resolve formaldehyde-induced RNA-protein crosslinks [82].
The fundamental challenge in atypical chain research lies in differentiating these rare modifications from their abundant counterparts and accurately characterizing their architecture amidst a complex background of similar chemical structures. This whitepaper addresses the key technical pitfalls in this differentiation process and provides detailed methodological guidance for researchers investigating the functions of K6, K27, and K29 ubiquitin linkages.
Quantitative Profiling of Atypical Ubiquitin Chains
Understanding the relative abundance and structural characteristics of atypical chains is fundamental to developing effective differentiation strategies. The following table summarizes key quantitative and structural data essential for experimental design.
Table 1: Quantitative Profiling and Characteristics of Atypical Ubiquitin Chains
| Linkage Type | Relative Abundance | Structural Features | Known Cellular Functions | Primary Technical Challenges |
|---|---|---|---|---|
| K6-linked | <0.5% of total ubiquitin pools [62] | Adopts compact conformation [27] | DNA damage response, mitophagy, resolution of RNA-protein crosslinks [82] | Extremely low abundance masks specific functions; antibody cross-reactivity |
| K27-linked | <0.5% of total ubiquitin pools [62] | Extended conformation with unique interfaces [27] | Cell proliferation, nuclear p97 activity, innate immune signaling [62] [33] | Poorly characterized recognition elements; low stoichiometry |
| K29-linked | ~1-2% of total ubiquitin pools [24] | Flexible chains with branching capability [83] | Chromosome biology, SUV39H1 degradation, H3K9me3 homeostasis [81] [62] | Branching complexity; discrimination from K48 linkages |
The table illustrates the significant abundance disparity between atypical and conventional ubiquitin chains, with all three atypical linkages representing minor ubiquitin populations. This quantitative disadvantage directly contributes to the primary technical challenges in their study. Furthermore, K29-linked chains demonstrate particular complexity through their capacity to form branched architectures with K48-linked chains, creating an additional layer of analytical difficulty [83].
Table 2: Enzymatic Machinery Governing Atypical Ubiquitin Chain Dynamics
| Linkage Type | E3 Ligases | Deubiquitinases (DUBs) | Chain Recognition Factors |
|---|---|---|---|
| K6-linked | BRCA1-BARD1, Parkin, RNF14 [82] [24] | USP11 (with UBL2 domain) [84] | p97/VCP, proteasome components [82] |
| K27-linked | TRIP12, RNF168, TRIM23 [62] [33] | TRABID, A20 [81] [33] | NEMO, Rhbdd3, p97 [62] [33] |
| K29-linked | Ufd4, TRIP12, HUWE1 [83] [24] | TRABID, USP11 [81] [84] | SUV39H1, proteasome receptors [81] [62] |
The enzymatic control of atypical chains involves specialized E3 ligases and DUBs that often demonstrate linkage preference rather than absolute specificity. This partial specificity creates significant challenges in pharmacological or genetic manipulation, as compensatory mechanisms frequently obscure phenotypic outcomes.
Methodological Pitfalls and Experimental Solutions
Linkage-Specific Detection and Antibody Limitations
The development and validation of linkage-specific reagents represents one of the most significant challenges in atypical chain research. Conventional antibodies raised against specific ubiquitin linkages often suffer from cross-reactivity issues that compromise data interpretation.
Pitfall: Antibody Cross-Reactivity Linkage-specific antibodies frequently demonstrate off-target binding to structurally similar chains or fail to recognize branched architectures containing their target linkage. For example, K29-linkages can form branched chains with K48-linkages [83], creating epitopes that may not be recognized by antibodies developed against homotypic K29 chains.
Experimental Solution: Orthogonal Validation
- Ubiquitin Replacement Strategy: Implement cell-based ubiquitin replacement systems enabling targeted conditional abrogation of individual ubiquitin linkages. This approach involves stable transfection of U2OS human osteosarcoma cells with doxycycline-inducible shRNAs targeting endogenous ubiquitin genes, rescued by expression of exogenous Ub harboring specific K-to-R mutations [62]. The system allows physiological validation of antibody specificity through linkage ablation.
- Tandem-Repeated Ubiquitin-Binding Entities (TUBEs): Employ TUBEs with linkage preference rather than absolute specificity, coupled with mass spectrometric validation. TUBEs can be engineered with varying affinities for different chain types and used in pull-down assays followed by middle-down MS analysis (Ub-clipping) to characterize captured chains [51].
Mass Spectrometric Characterization Challenges
Mass spectrometry has become the gold standard for ubiquitin chain characterization, but several pitfalls complicate the analysis of atypical chains.
Pitfall: Signal Suppression from Abundant Modifications The low stoichiometry of atypical chains means their diagnostic peptides are often suppressed during ionization by more abundant peptides from conventional ubiquitin chains or unmodified proteins.
Experimental Solution: Advanced Enrichment and Fragmentation Techniques
- DiGly Antibody Enrichment: Implement immunoaffinity enrichment using antibodies targeting the diglycine remnant (K-ε-GG) left after tryptic digestion of ubiquitinated proteins. Combine with linkage-specific antibodies for two-dimensional purification [51].
- Middle-Down MS Analysis (Ub-Clipping): Utilize the Ub-clipping method, which employs the viral DUB LbPro* to cleave ubiquitin chains after arginine 74, generating a C-terminal diGly remnant on the modified lysine. This allows direct mapping of branching points and linkage sites through detection of fragments with double-glycine remnants on both K29 and K48 residues [83].
- Cross-Linker Assisted Stabilization: For structural studies, employ chemical cross-linking strategies to stabilize enzymatic intermediates. For example, covalently link the catalytic cysteine of E3 ligases (e.g., Ufd4 C1450), the C-terminus of Ub, and the acceptor lysine (K29) of substrate to form a stable complex mimicking the transition state for cryo-EM analysis [83].
Genetic and Pharmacological Manipulation Issues
Pitfall: Compensatory Mechanisms and Redundancy Partial redundancy between ubiquitin acceptor lysines means that single linkage ablation may not produce phenotypic consequences due to compensatory chain formation through alternative lysines [24].
Experimental Solution: Multi-Layer Validation Framework
- Synthetic Genetic Array (SGA) Analysis: Conduct high-throughput genetic interaction screening by combining ubiquitin K-to-R mutants with gene deletion libraries. This approach identifies pathways regulated by specific linkage types through synthetic sick/lethal interactions [24].
- PROTAC Validation: Use proteolysis-targeting chimeras (PROTACs) to assess linkage requirements for targeted protein degradation. For example, proteasomal degradation is blocked specifically in Ub(K48R)-replaced cell lines when induced with a PROTAC driving VHL-dependent degradation of BRD4 [62].
- Enzyme Kinetics Profiling: Determine catalytic efficiency (kcat/Km) for E2/E3 combinations using defined ubiquitin chain substrates. For Ufd4, enzyme kinetics revealed ~5.2-fold higher efficiency for proximal K29 sites (0.11 μM⁻¹ min⁻¹) compared to distal K29 sites (0.021 μM⁻¹ min⁻¹) in K48-linked diUb substrates [83].
Visualizing Atypical Ubiquitin Pathways
The following diagrams illustrate key experimental workflows and signaling pathways involving atypical ubiquitin chains, providing visual guidance for methodological implementation.
Ubiquitin Replacement Strategy Workflow
K29/K48-Branched Ubiquitin Chain Formation
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents for Atypical Ubiquitin Chain Studies
| Reagent Category | Specific Examples | Function/Application | Technical Considerations |
|---|---|---|---|
| Ubiquitin Mutants | Ub(K29R), Ub(K27R), Ub(K6R) | Linkage ablation in replacement strategies | Structural perturbations minimal (in silico Si < 0.05) [62] |
| Linkage-Specific Antibodies | Anti-K29, Anti-K27, Anti-K6 | Immunoblot, immunofluorescence, enrichment | Require orthogonal validation; limited for branched chains |
| Specialized E2/E3 Pairs | Ubc4/Ufd4, UBCH5/TRIP12 | In vitro reconstitution of specific linkages | Context-dependent specificity; subcellular compartmentalization effects |
| Activity-Based Probes | K29/K48-branched triUb~probe~ | Trapping enzymatic intermediates for structural studies | Enable cryo-EM analysis of transition states [83] |
| DUB Inhibitors | Selective USP11 inhibitors (Fenoldopam, Olanzapine) | Probing K48 and K29 chain dynamics | FDA-approved drugs repurposed as chemical probes [84] |
| Mass Spec Standards | K29/K48-branched tetraUb | Method calibration and quantification | Enable detection of double-glycine remnants on both sites [83] |
The technical pitfalls in differentiating atypical ubiquitin chains from common modifications stem primarily from their low abundance, structural complexity, and overlapping enzymatic regulation. Success in this challenging field requires implementation of the multi-faceted approaches outlined in this technical guide: rigorous validation of linkage-specific reagents, advanced mass spectrometric methods with appropriate enrichment strategies, and genetic systems that account for compensatory mechanisms. The specialized functions of K6, K27, and K29 linkages in critical processes such as epigenome maintenance, DNA damage response, and immune signaling underscore the importance of overcoming these technical challenges. As methodological innovations continue to emerge, particularly in the areas of structural biology and proteomics, our capacity to decipher the complex biological functions of these atypical ubiquitin chains will undoubtedly expand, opening new avenues for therapeutic intervention in cancer, neurodegenerative diseases, and immune disorders.
Functional Validation and Comparative Analysis: Context-Dependent Signaling of Atypical Linkages
In the rigorous validation of gene function, particularly in the context of human disease modeling, phenotypic rescue experiments serve as a critical cornerstone. These experiments test a fundamental hypothesis: that reintroducing a wild-type gene or a specific variant into a diseased model organism can reverse or prevent a pathological phenotype. This methodology provides direct evidence of a gene's function and establishes a causal link between a genetic lesion and its cellular or organismal consequence. Within the complex landscape of post-translational modifications, research into the functions of atypical ubiquitin chain linkages—K6, K27, and K29—particularly benefits from this approach. The functional characterization of these linkages, which are abundant yet less understood than their K48 and K63 counterparts, often requires sophisticated genetic models and precise rescue paradigms to decode their specialized roles in cellular signaling, protein degradation, and homeostasis [19].
This whitepaper provides an in-depth technical guide for researchers and drug development professionals on designing, executing, and interpreting phenotypic rescue experiments. We place special emphasis on strategies relevant for probing the functions of atypical ubiquitin chains, a area with significant implications for understanding disease mechanisms and identifying new therapeutic targets.
Theoretical Foundations of Rescue Experiments
Core Principles and Definitions
A phenotypic rescue experiment is designed to demonstrate that the expression of a wild-type or engineered gene product can compensate for a genetic defect in a model system. A successful rescue provides strong evidence for the gene's necessity and sufficiency in producing the observed phenotype. The overarching logic follows a simple three-step process: (1) a genetic mutation is introduced, leading to a measurable phenotype; (2) the wild-type gene (or a functional variant) is reintroduced; and (3) the system is assessed for reversion to wild-type or near-wild-type conditions.
When applied to the study of ubiquitin linkages, the concept of "rescue" can be applied at multiple levels. For example, in a yeast strain engineered to express only a K11R mutant ubiquitin that cannot form K11-linked chains, a phenotype related to cell cycle progression might be rescued not only by re-introducing wild-type ubiquitin but also by expressing a engineered ubiquitin that can form a specific alternative chain type, thereby revealing functional redundancy or specificity [24].
The Framework for Model Organism Selection
Choosing an appropriate model organism is paramount. The selection should be guided by the biological question, the tools available for genetic manipulation, and the relevance of the model's biology to the human process being studied.
Challenging Assumptions in Model Selection: Recent frameworks advocate for a gene-centric rather than a purely phylogeny-based approach. Key assumptions to challenge include:
- Distant organisms can’t be useful models: Evolutionary proximity does not always guarantee functional similarity for a specific gene. A gene in a distant organism might behave more like the human version than its ortholog in a standard mammalian model [85].
- The model must mimic the human disease phenotype: A useful model may exhibit a different, yet mechanistically related, phenotype. A mutation in a ubiquitin-related gene might cause neurological defects in humans but a cell intercalation defect in a nematode, with both stemming from a common cytoskeletal failure [85].
- There is a single "best" model for every disease: A combination of organisms, each with complementary strengths (e.g., throughput, physiological similarity), is often more effective than a single model [85].
Table 1: Common Model Organisms for Rescue Studies in Ubiquitin Research
| Organism | Key Advantages | Limitations | Example Use Case |
|---|---|---|---|
| S. cerevisiae (Yeast) | Superior for genetics; high-throughput screening; well-characterized ubiquitin machinery [24] | Lack of complex tissues and some mammalian pathways | Genetic interactome mapping of ubiquitin lysine mutants [24] |
| C. elegans (Nematode) | Transparent body; invariant cell lineage; simple nervous system | Less relevant for organ-level diseases | |
| D. melanogaster (Fruit Fly) | Complex organ systems; powerful genetics; short generation time | Less relevant for some mammalian pathologies | |
| D. rerio (Zebrafish) | Vertebrate model; high fecundity; optical transparency for imaging; high-throughput CRISPR mutagenesis [86] | Lower throughput than invertebrates; not a mammal | Screening orthologs of human disease genes [86] |
| M. musculus (Mouse) | Mammalian physiology; sophisticated genetic tools; models of human disease | Expensive; slow; ethically complex |
Experimental Design and Workflows
A Generalized Workflow for Phenotypic Rescue
The following diagram outlines the core logical pathway for designing and interpreting a rescue experiment, from hypothesis generation to final conclusion.
Generating the Loss-of-Function Model
The first step is to create a model organism with a defined genetic lesion and a clear, measurable phenotype. CRISPR-Cas9 is the predominant technology for this purpose.
Detailed Protocol: CRISPR-Cas9 Mutagenesis in Vertebrates
- Design of guide RNAs (gRNAs): Design 2-3 gRNAs with high on-target efficiency and low off-target potential, targeting early exons of the gene of interest to maximize the chance of generating frameshifts and null alleles. For ubiquitin genes, which are often multi-copy, ensure gRNAs target all genomic copies or focus on specific chain-forming lysines via point mutations [86] [24].
- Microinjection in Zebrafish/Mice: Co-inject Cas9 mRNA or protein with synthesized sgRNAs into one-cell stage embryos. This induces double-strand breaks repaired by non-homologous end joining (NHEJ), leading to insertions or deletions (indels) [86].
- Validation of Mutants: Raise injected embryos (F0 founders) and screen for phenotypic manifestations. Outcross F0 fish to wild-types; a subset will transmit mutations to the F1 generation. Genotype F1 progeny to identify individuals with frameshift mutations. Establish stable mutant lines from these F1 carriers [86].
- Phenotypic Characterization: Conduct a detailed analysis of the F2 or F3 generation to establish the homozygous mutant phenotype. For ubiquitin-related genes, assays may include immunoblotting for substrate accumulation, cycloheximide chase assays for protein half-life, and microscopic analysis for developmental defects.
Rescue Strategies and Delivery Methods
The rescue construct can be delivered via several methods, each with advantages and considerations.
- DNA/RNA Microinjection: Synthetic mRNA or plasmid DNA encoding the wild-type gene can be injected into early-stage embryos. This is rapid and useful for assessing rescue of early developmental phenotypes but is transient and mosaic. mRNA is often preferred for its immediate translation and absence of integration concerns.
- Transgenesis: Creating a stable transgenic line carrying a rescue construct under a specific promoter ensures consistent, heritable expression. This is ideal for long-term studies and crossing into different genetic backgrounds. The use of Tol2 or other transposase systems in zebrafish facilitates genomic integration.
- Viral Transduction: In mammalian models, adeno-associated viruses (AAVs) or lentiviruses can be used to deliver rescue constructs to specific tissues postnatally, offering spatial and temporal control.
A key consideration for rescuing ubiquitin linkage function is the design of the rescue construct itself. For example, to test the specific function of a K27-linkage, one might rescue a ubiquitin-ligase knockout with a wild-type version of the E3 and a mutant version that is specifically defective in forming K27-linked chains but competent for other linkages [8].
Quantitative Phenotypic Assessment
Rescue must be quantified using robust, objective assays. The table below summarizes key quantitative metrics relevant to ubiquitin research.
Table 2: Quantitative Assays for Phenotypic Assessment in Ubiquitin Research
| Phenotypic Category | Measurable Readout | Technique | Data Analysis |
|---|---|---|---|
| Molecular Phenotype | Protein half-life, Substrate accumulation | Cycloheximide chase, Immunoblotting | Quantify band intensity; calculate half-life (t₁/₂) [19] |
| Substrate Ubiquitination | Abundance of specific ubiquitin linkages | Linkage-specific antibodies, Mass spectrometry | Spectral counting; intensity of K6/K27/K29 signals [19] [12] |
| Cellular Phenotype | Cell proliferation, Cell cycle arrest, Cell death | Flow cytometry (BrdU, PI, Annexin V) | Percentage of cells in each phase; fold-change vs. control |
| Organismal Phenotype | Survival, Morphological defects, Locomotor behavior | Kaplan-Meier curves, Imaging, Automated tracking | Hazard Ratio (HR); severity scoring; velocity/distance |
Application to Atypical Ubiquitin Chain Research
Signaling Pathways Involving Atypical Linkages
The following diagram integrates atypical ubiquitin chains into a key signaling pathway to illustrate potential nodes for rescue experiments.
Designing Rescue Constructs for Ubiquitin Studies
Rescuing pathways regulated by atypical ubiquitin chains requires precise construct design to establish linkage-specific function.
- Rescuing Ubiquitin Ligase Function: To prove that a specific E3 ligase acts through K27-linkages, one could rescue an E3-null model with:
- Wild-type E3: Should restore pathway function and K27-linked ubiquitination of substrates.
- Catalytic Dead E3 (C-to-A mutant): Should fail to rescue, confirming enzymatic activity is required.
- E3 Mutant Defective in K27-Specific E2 Binding: Should fail to restore K27-linked ubiquitination and pathway function, even if other E3 functions remain intact [8].
- Rescuing with Ubiquitin Mutants: In a model where all endogenous ubiquitin is mutated (e.g., K11R), phenotypes can be rescued with:
- Wild-type Ubiquitin: Should fully rescue.
- K11-only Ubiquitin (all other lysines mutated to Arg): Tests the sufficiency of K11-linkages for the process. This approach was used in yeast to reveal K11-linkages roles in cell cycle and amino acid import [24].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for Rescue Experiments in Ubiquitin Research
| Reagent / Tool | Function / Application | Example in Context |
|---|---|---|
| CRISPR-Cas9 System | Generation of knockout models and precise point mutations. | Introducing K-to-R mutations in endogenous ubiquitin loci in yeast or zebrafish [86] [24]. |
| Linkage-Specific Ubiquitin Antibodies | Detection of specific polyubiquitin chain topologies by immunoblotting or immunofluorescence. | Validating loss of K27-ubiquitination in a TRIM mutant and its restoration upon rescue [8]. |
| Mutant Ubiquitin Plasmids | Expression vectors for wild-type and lysine-deficient ubiquitin mutants (e.g., K6R, K27R, K29R, K11-only). | Testing the necessity and sufficiency of specific linkages in rescue assays [24] [19]. |
| Mass Spectrometry (Ubiquitin Profiling) | Proteome-wide identification and quantification of ubiquitination sites and linkage types. | Discovering novel substrates and confirming the specific linkage types altered in the mutant and restored during rescue [19]. |
| Base & Prime Editors | Introduction of precise point mutations without double-strand breaks, minimizing indel formation. | Creating clean, patient-specific point mutations in model organisms for precise rescue studies [86]. |
| Proteasome Inhibitors | Block degradation of proteasome-targeted proteins, allowing accumulation of ubiquitinated substrates. | Stabilizing K48/K11-branched ubiquitin chains on APC/C substrates to study their rescue dynamics [12]. |
Phenotypic rescue remains the gold-standard experiment for establishing a causal relationship between a gene and its function in a physiological context. When applied to the complex field of atypical ubiquitin chains, these experiments require meticulous design, including the use of appropriate and sometimes unconventional model organisms, precise genetic editing with CRISPR, and the engineering of sophisticated rescue constructs that can test linkage-specific hypotheses. The quantitative frameworks and tools outlined in this guide provide a pathway for researchers to definitively characterize the roles of K6, K27, K29, and other ubiquitin linkages in health and disease. As drug discovery increasingly targets the ubiquitin-proteasome system, the ability to rigorously validate the function of these pathways in vivo through rescue experiments will be instrumental in translating basic research into novel therapeutics.
Ubiquitination is a crucial post-translational modification that regulates a vast array of cellular processes in eukaryotes. The versatility of ubiquitin signaling stems from the ability of this 76-amino acid protein to form diverse polymeric chains through its seven internal lysine residues (K6, K11, K27, K29, K33, K48, K63) or its N-terminal methionine (M1) [27]. For decades, research has predominantly focused on the canonical K48-linked chains, which target substrates for proteasomal degradation, and K63-linked chains, which regulate non-proteolytic signaling pathways. However, emerging evidence reveals that atypical ubiquitin chains linked through K6, K27, and K29 serve specialized biological functions that expand the coding potential of the ubiquitin system [87] [12]. These non-canonical linkages account for a smaller percentage of cellular ubiquitin chains but play critical roles in specific physiological and pathological contexts, including immune regulation, protein quality control, and mitochondrial homeostasis. This technical guide provides a comprehensive comparison of the structural features, functional properties, and experimental methodologies for studying these atypical ubiquitin chains in relation to their canonical counterparts.
Structural and Functional Characteristics by Linkage Type
Canonical Ubiquitin Chains
Table 1: Properties of Canonical Ubiquitin Chains
| Linkage Type | Abundance | Primary Functions | Chain Conformation | Key E3 Ligases | Associated DUBs |
|---|---|---|---|---|---|
| K48 | ~30% in yeast [24] | Proteasomal degradation [24] | Closed conformation [24] | HUWE1 [88] | OTUB1 [14], USP14 [89] |
| K63 | Less abundant than K48 [24] | DNA repair, NF-κB signaling, inflammation, protein trafficking [7] [24] | Extended conformation [24] | TRAF6 [88] | AMSH [14], CYLD [88] |
K48-linked chains represent the most abundant ubiquitin linkage type and serve as the primary signal for targeting proteins to the 26S proteasome for degradation [24]. These chains adopt a compact structure with the hydrophobic patches of adjacent ubiquitin monomers engaged in inter-domain contacts, facilitating recognition by proteasomal receptors [24]. In contrast, K63-linked chains assume a more extended conformation with minimal non-covalent interactions between ubiquitin units, making them suitable for their roles in assembly of signaling complexes, DNA damage response, endocytic trafficking, and inflammatory pathway activation [7] [24].
Atypical Ubiquitin Chains
Table 2: Properties of Atypical Ubiquitin Chains
| Linkage Type | Abundance | Primary Functions | Structural Features | Key E3 Ligases | DUB Sensitivity |
|---|---|---|---|---|---|
| K6 | Low [24] | DNA damage response, mitophagy [14] [24] | Unknown | BRCA1-BARD1, Parkin [24] | Variable DUB sensitivity [14] |
| K27 | Low [24] | Mitophagy, antiviral innate immunity [87] [14] | No noncovalent interdomain contacts [14] | Parkin [24], mitochondrial E3s | Resistant to most DUBs [14] |
| K29 | Low [24] | Wnt signaling, mRNA stability, proteostasis [14] [24] | Weak/transient interdomain contacts [14] | Ufd4, UBE3C [12] [24] | Resistant to Ubp6 [14] |
K6-linked chains have been implicated in the DNA damage response through the BRCA1-BARD1 E3 ligase complex and in mitophagy through Parkin-mediated ubiquitination of mitochondrial proteins [24]. These chains appear to function in a proteolysis-independent manner in DNA repair, while their role in mitophagy may involve regulating protein stability or interactions.
K27-linked chains exhibit unique biochemical properties, including remarkable resistance to deubiquitination by most deubiquitinating enzymes (DUBs) [14]. This linkage remains uncleaved by linkage-nonspecific DUBs including USP2, USP5, and Ubp6, which effectively cleave other ubiquitin linkages [14]. Structurally, K27-Ub2 shows no evidence of noncovalent interdomain contacts in the distal ubiquitin unit, while the proximal ubiquitin displays significant chemical shift perturbations [14]. Functionally, K27 linkages play important roles in mitochondrial quality control by decorating proteins such as Miro1 to regulate mitophagy and serve as markers of mitochondrial damage [14]. Additionally, K27-linked chains contribute to the regulation of antiviral innate immune signaling pathways [87].
K29-linked chains participate in diverse cellular processes including Wnt/β-catenin signaling, regulation of mRNA stability through the adaptor protein UBXD8, and protein quality control pathways [14] [24]. Similar to K27 linkages, K29-Ub2 shows resistance to certain DUBs like Ubp6 and exhibits weak or transient interdomain contacts [14]. In yeast, the E3 ligases Ufd4 and Ufd2 collaborate to synthesize branched K29/K48 chains on substrates of the ubiquitin fusion degradation (UFD) pathway [12].
Branched Ubiquitin Chains and Complex Topologies
Beyond homotypic chains, ubiquitin can form heterotypic structures including mixed linkage chains and branched chains where a single ubiquitin molecule is modified at multiple lysine residues. Branched ubiquitin chains account for 10-20% of total ubiquitin polymers and exhibit specialized functional properties [89].
Table 3: Characteristics of Branched Ubiquitin Chains
| Branched Chain Type | Biological Function | Synthesis Mechanism | Functional Outcome |
|---|---|---|---|
| K48-K63 [88] | NF-κB signaling amplification | HUWE1 adds K48 branches to TRAF6-synthesized K63 chains [88] | Protects K63 chains from CYLD-mediated cleavage [88] |
| K11-K48 [89] | Cell cycle progression, proteotoxic stress response | APC/C with UBE2C and UBE2S; UBR5 [89] [12] | Accelerated proteasomal degradation [89] |
| K29-K48 [12] | Ubiquitin fusion degradation pathway | Ufd4 (K29) and Ufd2 (K48) collaboration [12] | Substrate degradation [12] |
The K48-K63 branched ubiquitin chain represents a particularly well-characterized example that plays a crucial role in NF-κB signaling regulation [88]. In response to interleukin-1β stimulation, the E3 ligase HUWE1 generates K48 branches on K63-linked chains assembled by TRAF6 [88]. These branched linkages are recognized by the TAB2 complex but are protected from deubiquitination by CYLD, thereby amplifying NF-κB activation signals [88].
K11-K48 branched chains serve as priority signals for proteasomal degradation, facilitating the timely turnover of cell cycle regulators and misfolded proteins [89]. Structural studies reveal that the human 26S proteasome recognizes K11/K48-branched ubiquitin chains through a multivalent mechanism involving RPN2 and RPN10, with the proteasome-associated DUB UCHL5 showing preference for processing these branched chains [89].
Experimental Methods for Studying Ubiquitin Linkages
Linkage-Specific Detection and Analysis
Tandem Ubiquitin Binding Entities (TUBEs): TUBEs are engineered affinity reagents composed of multiple ubiquitin-associated (UBA) domains that exhibit nanomolar affinity for polyubiquitin chains [7]. Linkage-specific TUBEs have been developed that can differentiate between K48- and K63-linked ubiquitination in high-throughput assays [7] [52]. These tools enable researchers to capture and quantify endogenous protein ubiquitination in a linkage-specific manner without requiring genetic manipulation of the ubiquitin system. For example, K63-TUBEs specifically capture RIPK2 ubiquitination induced by L18-MDP stimulation, while K48-TUBEs capture PROTAC-induced RIPK2 ubiquitination [7].
Mass Spectrometry-Based Approaches: Absolute quantification (AQUA) of ubiquitin linkages using mass spectrometry provides a comprehensive method for profiling chain topology [88] [89]. This approach involves spiking samples with stable isotope-labeled internal standards corresponding to specific ubiquitin linkage peptides, enabling precise quantification of chain types present in complex biological samples [88]. Additionally, Ub-AQUA can identify branched chains by detecting doubly ubiquitinated peptides [89].
Genetic Approaches: Systematic genetic interaction screening in yeast has revealed pathways regulated by specific ubiquitin linkages [24]. By combining gene deletions with ubiquitin mutants that abrogate specific linkages (lysine-to-arginine mutations), researchers have identified synthetic genetic interactions that uncover functional roles for atypical chains. For example, K11R ubiquitin mutants show strong genetic interactions with threonine biosynthetic genes and components of the anaphase-promoting complex [24].
Specialized Research Reagents
Table 4: Essential Research Reagents for Ubiquitin Chain Analysis
| Reagent/Tool | Composition/Type | Specificity | Key Applications |
|---|---|---|---|
| K63-TUBE [7] [52] | Tandem ubiquitin-binding entities | K63-linked chains | Capturing inflammatory signaling ubiquitination |
| K48-TUBE [7] [52] | Tandem ubiquitin-binding entities | K48-linked chains | Detecting degradative ubiquitination |
| Linkage-specific DUBs [14] | Deubiquitinating enzymes | Various linkages | Linkage identification and cleavage validation |
| Ubiquitin mutants (K-to-R) [24] | Mutant ubiquitin alleles | Specific linkage ablation | Genetic interaction studies |
| Linkage-specific antibodies [89] | Monoclonal/polyclonal antibodies | Specific linkage types | Immunoblotting and immunohistochemistry |
Diagrams of Ubiquitin Signaling Pathways
K48-K63 Branched Ubiquitin in NF-κB Signaling
K11/K48 Branched Chain Recognition by Proteasome
Experimental Workflow for Linkage-Specific Analysis
Research Applications and Therapeutic Implications
The specialized functions of atypical ubiquitin chains present intriguing possibilities for therapeutic intervention. K63-linked ubiquitination has emerged as a promising target for modulating inflammatory responses, with inhibitors targeting enzymes like TRAF6, Ubc13, and Mms2 showing potential in preclinical models of rheumatoid arthritis and colitis [7]. Additionally, PROTAC (Proteolysis Targeting Chimeras) technology leverages the K48 ubiquitination pathway to target disease-relevant proteins for degradation, with several candidates advancing through clinical trials [7] [90].
The unique properties of atypical chains also offer therapeutic opportunities. The resistance of K27-linked chains to most DUBs suggests potential strategies for stabilizing ubiquitin signals on specific targets [14]. Furthermore, the priority degradation signal provided by K11/K48-branched chains could be harnessed to enhance the efficiency of targeted protein degradation approaches [89]. Understanding the distinct roles of these ubiquitin linkages in cancer hallmarks, including metabolic reprogramming, immune evasion, and cellular plasticity, may reveal new vulnerabilities for therapeutic exploitation [90].
The expanding landscape of atypical ubiquitin chains linked through K6, K27, and K29 represents a sophisticated extension of the ubiquitin code that enables precise regulation of specialized cellular processes. While canonical K48 and K63 chains govern fundamental pathways of protein degradation and signal transduction, the atypical linkages provide nuanced control over mitochondrial function, immune responses, and protein quality control mechanisms. Advanced methodological approaches including linkage-specific TUBEs, quantitative mass spectrometry, and genetic interaction mapping have begun to decode the functions of these non-canonical ubiquitin signals. The integration of these tools with structural insights and mechanistic studies continues to illuminate the complex interplay between different ubiquitin chain types in health and disease, offering exciting prospects for developing novel therapeutic strategies that target specific aspects of the ubiquitin system.
The ubiquitin code, once thought to be primarily mediated through K48 and K63-linked chains, has expanded to include atypical ubiquitin linkages (K6, K11, K27, K29, K33, and M1/linear) that play sophisticated roles in cellular signaling networks. Unlike their canonical counterparts, atypical chains frequently operate through collaborative mechanisms, forming homotypic, mixed-linkage, and branched structures that enable signal integration and fine-tuning of cellular processes. This technical review examines the molecular architecture of atypical ubiquitin chain signaling cross-talk, focusing on their integrated functions in antiviral immunity, cell cycle regulation, and inflammatory pathways. We provide comprehensive analysis of the E3 ligase partnerships that assemble complex ubiquitin architectures, detail experimental methodologies for decoding these networks, and present resource tools for continued investigation. The emerging paradigm reveals that atypical chains function not in isolation but as interconnected components of a sophisticated ubiquitin signaling network that processes biological information through collaborative chain assembly and recognition.
Ubiquitin, a 76-amino acid protein, regulates virtually all cellular processes through post-translational modification of target proteins. The conventional view distinguished primarily between K48-linked chains (targeting proteins for proteasomal degradation) and K63-linked chains (non-degradative signaling). However, advancing methodologies have revealed that atypical ubiquitin chains—those linked through K6, K11, K27, K29, K33, or the N-terminal methionine (M1)—comprise a sophisticated regulatory layer that enables complex information processing in cells [80] [91].
These atypical linkages create a diverse ubiquitin vocabulary that expands the signaling capacity of this modification. While historically challenging to study due to technical limitations and their relatively low abundance compared to canonical chains, atypical linkages are now recognized as critical regulators in specific biological contexts, particularly through their ability to collaborate with other ubiquitin chains and post-translational modifications [8] [92]. This cross-talk enables precise control over processes ranging from innate immune activation to cell cycle progression and apoptotic regulation.
The following sections examine the architectural principles governing atypical chain collaboration, detail their functional integration in specific signaling pathways, present experimental approaches for their study, and provide key resources for researchers investigating this expanding field of ubiquitin signaling.
Chain Architecture and Collaborative Assembly
Atypical ubiquitin chains exhibit diverse structural configurations that define their functional capabilities and collaborative potential. The classification system organizes these chains into distinct architectural categories, each with unique properties that influence their signaling capacity.
Figure 1: Architectural diversity of atypical ubiquitin chains. Chains are classified by their linkage patterns, with branched configurations enabling particularly complex signaling integration.
Classification of Atypical Ubiquitin Chains
- Homotypic chains: Uniform chains using the same lysine residue throughout (e.g., all K27 linkages) [80]
- Mixed-linkage chains: Chains utilizing different lysines in sequence without branching [80]
- Branched chains: Ubiquitin monomers simultaneously modified on at least two different acceptor sites, creating fork-like structures [12]
- Heterologous chains: Integration of ubiquitin-like modifiers (SUMO, NEDD8) into ubiquitin chains [80]
Mechanisms of Collaborative Chain Assembly
The synthesis of complex ubiquitin architectures frequently requires coordinated action between multiple E3 ligases with distinct linkage specificities. This collaborative assembly enables temporal control over signaling outcomes and creates information-rich ubiquitin signatures.
E3 Ligase Partnerships represent a fundamental mechanism for branched chain assembly. During NF-κB signaling, TRAF6 (synthesizing K63-linked chains) collaborates with HUWE1 (adding K48 linkages) to create branched K48/K63 chains that modulate signal amplitude and duration [12]. Similarly, in apoptosis regulation, ITCH and UBR5 cooperate to convert non-proteolytic K63-linked chains on TXNIP to degradative branched K48/K63 chains, effectively switching signaling from activation to termination [12].
Sequential Assembly Pathways provide another mechanism for collaboration. The anaphase-promoting complex/cyclosome (APC/C) cooperates with two different E2 enzymes (UBE2C and UBE2S) to form branched K11/K48 chains during mitosis. UBE2C first attaches short chains containing mixed linkages, then UBE2S adds multiple K11 linkages to create branched polymers that enhance substrate recognition and degradation efficiency [12] [24].
Table 1: E3 Ligase Partnerships in Branched Chain Synthesis
| Branched Chain Type | Collaborating E3 Ligases | Biological Process | Functional Outcome |
|---|---|---|---|
| K48/K63 | TRAF6 + HUWE1 | NF-κB signaling | Signal modulation [12] |
| K48/K63 | ITCH + UBR5 | Apoptosis regulation | Signal termination [12] |
| K29/K48 | Ufd4 + Ufd2 | Ubiquitin fusion degradation | Proteosomal targeting [12] |
| K11/K48 | APC/C (UBE2C + UBE2S) | Mitotic progression | Cell cycle regulation [12] [24] |
Functional Integration in Signaling Pathways
Atypical ubiquitin chains serve as critical integration points in complex signaling networks, particularly in antiviral immunity and cell cycle control. Their ability to collaborate with other ubiquitin linkages enables precise regulation of pathway activation, duration, and termination.
Antiviral Innate Immune Signaling
The antiviral response demonstrates sophisticated collaboration between atypical and canonical ubiquitin chains that determines signaling outcomes through RIG-I-like receptors (RLRs) and DNA sensing pathways.
Figure 2: Atypical ubiquitin chains in antiviral innate immune signaling. Multiple atypical linkages collaborate to regulate transcription factor activation and immune output determination.
K27-linked chains serve both activating and regulatory roles in RLR signaling. TRIM23 conjugates K27-linked chains to NEMO, promoting activation of both NF-κB and IRF3 transcription factors [8] [33]. Conversely, K27-linked ubiquitination also recruits negative regulators; Rhbdd3 binds K27 chains on NEMO and recruits the deubiquitinase A20, which removes K63-linked chains to prevent excessive NF-κB activation [8] [33]. This collaboration between K27 and K63 linkages enables precise signal calibration.
Linear (M1-linked) chains demonstrate pathway-specific regulation. LUBAC-mediated linear ubiquitination of NEMO potentiates NF-κB signaling through high-affinity binding to the UBAN domain of NEMO [8] [33]. Simultaneously, linear chains disrupt the MAVS-TRAF3 interaction, inhibiting IRF3 activation and type I interferon production while promoting NF-κB-driven inflammatory cytokine production [8]. This collaborative outcome directs the immune response toward specific functional outputs.
K11-linked chains primarily regulate protein stability in immune signaling. RNF26-mediated K11-linked ubiquitination of STING inhibits its degradation, potentiating type I interferon production [8]. Meanwhile, K11/K48-branched chains on Beclin-1 promote its proteasomal degradation, limiting Beclin-1-mediated disruption of RIG-I/MAVS interaction and thereby enhancing interferon production [8]. This demonstrates collaboration between K11 and K48 linkages in stability regulation.
Table 2: Atypical Ubiquitin Chain Functions in Antiviral Immunity
| Linkage Type | E3 Ligase(s) | Substrate(s) | Functional Outcome | Collaborative Partners |
|---|---|---|---|---|
| Linear (M1) | LUBAC | NEMO, MAVS | NF-κB activation, IFN inhibition [8] | K63 chains (competes for NEMO binding) |
| K11 | RNF26 | STING, IRF3 | STING stabilization, IRF3 degradation [8] | K48 chains (branched degradation signals) |
| K27 | TRIM23, TRIM21 | NEMO, MAVS | NF-κB and IRF3 activation [8] | K63 chains (A20-mediated removal) |
| K27/K29 | RNF34 | MAVS | Autophagic degradation, IFN restriction [8] | Selective autophagy receptors |
| K29 | SKP1-Cullin-Fbx21 | ASK1 | IFNβ and IL-6 production [8] | MAPK signaling components |
Cell Cycle Regulation and Protein Degradation
Beyond immune signaling, atypical chains collaborate to control cell cycle progression and protein degradation. The anaphase-promoting complex/cyclosome (APC/C) utilizes branched K11/K48 chains to ensure efficient substrate recognition and degradation during mitotic exit [24]. Genetic studies in Saccharomyces cerevisiae reveal that K11-linkage mutants exhibit strong genetic interactions with APC/C components, indicating functional collaboration between K11 and K48 linkages in cell cycle regulation [24].
The collaboration between atypical chains in degradation signals represents an important regulatory mechanism. Branched K11/K48 chains appear to enhance degradation efficiency compared to homotypic K48 chains alone, potentially through improved proteasome recognition or binding avidity [12]. Similarly, K29/K48-branched chains serve as specialized degradation signals in the ubiquitin fusion degradation (UFD) pathway, demonstrating how atypical linkages can create specialized degradation signals for specific cellular pathways [12].
Experimental Approaches and Methodologies
Decoding the collaborative functions of atypical ubiquitin chains requires specialized methodologies that can distinguish between linkage types, identify branched structures, and determine functional consequences.
Genetic Interaction Mapping
Systematic genetic interaction analysis has proven powerful for identifying pathways regulated by specific ubiquitin linkages. In one comprehensive approach, yeast strains expressing lysine-to-arginine ubiquitin mutants (incapable of forming specific linkages) were crossed with a gene deletion library to identify synthetic genetic interactions [24]. This method revealed that K11R mutants had strong genetic interactions with threonine biosynthetic genes and APC/C components, uncovering previously unknown roles for K11 linkages in amino acid import and cell cycle regulation [24].
Protocol: Synthetic Genetic Array (SGA) with Ubiquitin Mutants
- Engineer yeast strains expressing K-to-R ubiquitin mutants at all endogenous ubiquitin loci
- Verify near-wild-type ubiquitin expression levels via immunoblotting
- Mate ubiquitin mutant strains with arrayed gene deletion library
- Induce sporulation and select for haploid double mutants
- Quantify colony growth phenotypes to identify genetic interactions
- Validate specific interactions through manual crosses and secondary assays
This approach identified thousands of candidate genetic interactions, providing a global view of pathways dependent on specific ubiquitin linkages and revealing their collaborative relationships with cellular processes [24].
Biochemical Reconstitution of Branched Chain Assembly
In vitro reconstitution of branched ubiquitin chain synthesis allows precise determination of the enzymatic requirements and mechanisms of collaborative E3 function.
Protocol: Branched Chain Assembly and Analysis
E3 Partnership Identification:
- Co-immunoprecipitation of candidate E3 pairs from stimulated cells
- Reciprocal immunoprecipitation to verify interactions
- Knockdown/rescue experiments to confirm functional collaboration
In Vitro Ubiquitination Assay:
- Purify E1, relevant E2s, and collaborating E3 ligases
- Set up reactions with ubiquitin, ATP-regenerating system
- Include specific E2 enzyme combinations (e.g., UBE2C + UBE2S for APC/C)
- Analyze products by SDS-PAGE and immunoblotting with linkage-specific antibodies
Chain Architecture Determination:
- Use ubiquitin mutants (K-to-R) to restrict available linkage sites
- Express defined ubiquitin chains as primers for extension
- Apply tandem ubiquitin-binding entities (TUBEs) to purify chains
- Analyze by mass spectrometry to identify branching points
This approach confirmed that UBR5 recognizes K63-linked chains synthesized by ITCH through its UBA domain, then adds K48 linkages to create branched K48/K63 chains on TXNIP [12].
Linkage-Specific Proteomic Analysis
Advancements in mass spectrometry and affinity reagents have enabled proteomic identification of atypical ubiquitination sites and chain architectures.
Protocol: Affinity Purification and Ubiquitin Remnant Profiling
Sample Preparation:
- Express His-tagged ubiquitin or use di-glycine remnant capture
- Treat cells with proteasome inhibitor (MG132) to accumulate ubiquitinated proteins
- Denature lysates to disrupt non-covalent interactions
Enrichment of Ubiquitinated Proteins:
- Immobilized metal affinity chromatography (IMAC) for His-tagged ubiquitin
- Antibody-based enrichment for di-glycine remnants
- TUBE domains for broad ubiquitin chain capture
Mass Spectrometry Analysis:
- Trypsin digestion generates di-glycine remnant signature on modified lysines
- LC-MS/MS with fragmentation to identify modified peptides
- Use linkage-specific antibodies to enrich for particular chain types
- Data analysis with software capable of identifying branched chains
This methodology has identified numerous proteins modified with atypical ubiquitin chains and revealed the prevalence of branched chains in cellular regulation [12].
Research Reagent Solutions
Investigating atypical ubiquitin chain collaboration requires specialized reagents designed to detect, manipulate, and analyze these modifications. The following toolkit provides essential resources for researchers in this field.
Table 3: Essential Research Reagents for Atypical Ubiquitin Chain Studies
| Reagent Category | Specific Examples | Research Application | Key Features & Considerations |
|---|---|---|---|
| Linkage-Specific Antibodies | Anti-K27-linkage, Anti-K11-linkage, Anti-linear linkage | Immunoblotting, immunofluorescence, immunoprecipitation | Variable specificity; validate with ubiquitin mutants; critical for in vivo assessment |
| Ubiquitin Mutants | K-to-R mutants (K11R, K27R, etc.), K-only mutants | Genetic studies, biochemical reconstitution | K48R requires wild-type ubiquitin co-expression for viability; essential for linkage assignment |
| Activity-Based Probes | Ubiquitin vinyl sulfones, HA-Ub-VS | Deubiquitinase specificity profiling | Identify DUBs with selectivity for atypical linkages; mechanistically informative |
| Affinity Capture Reagents | TUBEs (Tandem Ubiquitin Binding Entities), linkage-specific UBDs | Enrichment of polyubiquitinated proteins | TUBEs capture various chain types; linkage-specific UBDs offer selectivity |
| Branched Chain Standards | Chemically synthesized K11/K48-branched di-ubiquitin | Mass spectrometry reference, biochemical assays | Enable development of detection methods; quantify branched chains |
| E3 Ligase Inhibitors | LUBAC inhibitor HOIPIN-8, HECT domain inhibitors | Functional perturbation studies | Limited availability for atypical linkage-specific E3s; genetic approaches often necessary |
The study of atypical ubiquitin chains has evolved from identifying novel linkage types to understanding their collaborative functions in integrated signaling networks. The emerging paradigm reveals that atypical chains rarely operate in isolation but instead form collaborative partnerships with canonical chains and other post-translational modifications to create a sophisticated regulatory language. Branched chains, in particular, represent a sophisticated mechanism for signal integration, enabling conversion between chain types and creating unique recognition surfaces for downstream effectors.
Future research directions will need to address several key challenges: developing more specific tools for manipulating and detecting specific atypical linkages in cells, understanding the structural basis for branched chain recognition by ubiquitin-binding domains, and elucidating how collaborative ubiquitin signals are interpreted in physiological and pathological contexts. As our methodological capabilities advance, the continuing deciphering of the collaborative ubiquitin code will undoubtedly reveal new regulatory mechanisms and therapeutic opportunities across human diseases.
The ubiquitin code, a pivotal post-translational regulatory system, achieves remarkable functional diversity through the assembly of polyubiquitin chains connected via specific lysine residues. While the roles of canonical linkages like K48 and K63 are well-established, the functions of atypical linkages (K6, K27, K29) have remained more elusive. This whitepaper synthesizes recent structural and biochemical advances that illuminate the pathway-specific roles of these atypical ubiquitin chain types. We detail their specialized functions in innate immune signaling, proteostasis regulation, and cell cycle control, providing experimental frameworks and resource tools to advance research in this evolving field. The emerging paradigm reveals that these linkages constitute specialized components of the ubiquitin code with unique structural properties and recognition characteristics that dictate specific cellular outcomes.
Ubiquitination involves the covalent attachment of the small protein ubiquitin to substrate proteins, a process catalyzed by sequential E1, E2, and E3 enzyme activities [27]. The versatility of ubiquitin signaling stems from its capacity to form polyubiquitin chains through eight different linkage sites (K6, K11, K27, K29, K33, K48, K63, and M1). Atypical ubiquitin linkages (K6, K27, K29) are characterized by their low abundance under normal cycling conditions and their association with specialized cellular functions beyond proteasomal degradation [62]. Recent methodological advances have enabled detailed characterization of these linkages, revealing their critical contributions to cellular regulation. This technical guide integrates current understanding of their pathway-specific functions, with particular emphasis on innate immunity, proteostasis maintenance, and cell cycle control.
Functional Landscape of Atypical Ubiquitin Linkages
Table 1: Functional Roles of Atypical Ubiquitin Linkages in Key Cellular Pathways
| Linkage | Innate Immunity | Proteostasis | Cell Cycle | Structural Features | Key Enzymes |
|---|---|---|---|---|---|
| K6 | DNA damage signaling [62] | RNA-protein crosslink resolution [62] | - | Adopts extended conformation; recognized by TAB2 NZF domain [93] | RNF14 [62], Parkin (branched chains) [12] |
| K27 | Mitochondrial immunity regulation [14] [94] | Resistance to most DUBs [14] | Essential for cell proliferation [62] | Minimal noncovalent interdomain contacts [14] | RNF168 [62] |
| K29 | - | Proteotoxic stress response [62] | Wnt/β-catenin signaling [95] | Forms branched chains with K48 linkages [96] | TRIP12 [96] [62], UBE3C [12] |
Cellular Abundance and Functional Significance
Under normal cycling conditions, atypical ubiquitin linkages K6, K27, and K33 are present in low abundance in mammalian cells (typically <0.5% of total ubiquitin chains) [62]. Despite their low abundance, genetic studies using ubiquitin replacement cell lines have demonstrated that K27 linkages are indispensable for cell proliferation, whereas K6, K29, and K33 linkages are dispensable under standard conditions [62]. This suggests that K27 linkages may regulate essential housekeeping functions, while other atypical linkages likely serve more specialized roles in stress adaptation and pathway regulation.
Pathway-Specific Roles and Molecular Mechanisms
Innate Immune Signaling
Atypical ubiquitin linkages play specialized roles in fine-tuning innate immune responses, particularly in mitochondrial immunity and DNA damage signaling:
K27 Linkages in Mitochondrial Immunity: K27-linked ubiquitin chains have been implicated in regulating innate immune responses, potentially through their presence on mitochondrial proteins [14] [94]. Mitochondria, as remnants of engulfed α-proteobacteria, harbor molecules that can elicit innate immune responses if accumulated in the cytosol. K27-linked ubiquitination of mitochondrial proteins like Miro1 may serve as a marker of mitochondrial damage and participate in immune signaling pathways [14] [94].
K6 Linkages in DNA Damage Repair: K6-linked chains are involved in DNA damage signaling, with RNF168 generating K27-linked ubiquitin chains at DNA double-strand break sites that provide a scaffold for the recruitment of downstream repair factors [62]. This function creates a specialized signaling platform that coordinates the DNA damage response.
Diagram: K27 ubiquitin linkage in mitochondrial innate immunity
Proteostasis Regulation
The maintenance of protein homeostasis (proteostasis) involves multiple specialized functions of atypical ubiquitin linkages:
K29 Linkages in Proteotoxic Stress: K29-linked ubiquitin chains are heavily upregulated during proteotoxic stress, where they colocalize with stress granule components and enhance degradation signaling by facilitating p97/VCP-mediated unfolding of substrates [62]. This function is particularly important for extracting degradation substrates embedded in macromolecular structures or membranes.
K27 Linkages and DUB Resistance: K27-Ub2 exhibits unique biochemical properties, including resistance to cleavage by most deubiquitinases (DUBs) [14]. Screening against multiple DUB families (Cezanne, OTUB1, AMSH, USP2, USP5, and Ubp6) revealed that K27-Ub2 resisted cleavage by linkage non-specific DUBs including USP2, USP5, and Ubp6, with K27 being the only linkage that resisted cleavage by USP5 (IsoT) [14]. This property may contribute to the stability of K27-linked ubiquitin signals in proteostasis pathways.
K6 Linkages in Quality Control: K6-linked ubiquitin chains driven by RNF14 have been implicated in proteasome- and p97-dependent resolution of RNA-protein crosslinks, representing a specialized proteostasis mechanism [62].
Diagram: K29 ubiquitin linkage in proteotoxic stress response
Cell Cycle Control and Signaling
Atypical ubiquitin linkages participate in critical cell cycle regulatory pathways:
K29 Linkages in Wnt/β-catenin Signaling: K29-polyubiquitination participates in growth and development-associated pathways, including Wnt/β-catenin signaling [95]. This pathway is crucial for cell fate determination and is frequently dysregulated in cancer, placing K29 linkages in a key regulatory position for cell cycle progression and differentiation decisions.
K27 Linkages in Cell Proliferation: Genetic evidence from ubiquitin replacement studies has demonstrated that K27 linkages are essential for cell proliferation, though the precise mechanisms remain under investigation [62]. This suggests that K27 linkages regulate fundamental processes required for cell division.
Branched K11/K48 Chains in Mitotic Regulation: The APC/C (Anaphase-Promoting Complex/Cyclosome) cooperates with two different E2 enzymes (UBE2C and UBE2S) to form branched K11/K48 chains on substrates during mitosis [12]. These branched chains represent a specialized ubiquitin signal that ensures precise timing of mitotic progression.
Experimental Approaches and Methodologies
Determining Ubiquitin Chain Linkage
Table 2: Experimental Protocol for Ubiquitin Linkage Determination
| Step | Reagents | Concentrations | Purpose | Key Controls |
|---|---|---|---|---|
| Reaction Setup | E1 Enzyme, E2 Enzyme, E3 Ligase, Ubiquitin mutants, MgATP, 10X E3 Reaction Buffer | E1: 100 nM, E2: 1 μM, E3: 1 μM, Ubiquitin: ~100 μM | Establish in vitro ubiquitination system | Negative control: replace MgATP with dH₂O |
| K-to-R Mutant Screening | Seven Ubiquitin K-to-R mutants (K6R, K11R, K27R, K29R, K33R, K48R, K63R) | Each mutant: ~100 μM | Identify required lysine for chain linkage | Wild-type ubiquitin reaction |
| K-Only Mutant Verification | Seven Ubiquitin K-Only mutants (K6-only, K11-only, etc.) | Each mutant: ~100 μM | Verify specific linkage capability | Wild-type ubiquitin reaction |
| Analysis | SDS-PAGE, Western blot, anti-Ubiquitin antibody | Standard concentrations | Visualize ubiquitin chain formation | Include molecular weight markers |
The protocol for determining ubiquitin chain linkage employs a systematic approach using ubiquitin mutants to identify specific linkage requirements [97]. When all K-to-R mutants still form chains, this suggests either M1 (linear) linkage or mixed/branched chains containing multiple linkages [97].
Structural Characterization Techniques
NMR Spectroscopy: Solution NMR spectroscopy provides atom-specific information for each ubiquitin unit within chains, enabling quantification of amide chemical shift perturbations (CSPs) that indicate noncovalent interdomain contacts and structural perturbations [14].
Small-Angle Neutron Scattering (SANS): SANS combined with computational ensemble modeling reveals solution-state structures and dynamics of ubiquitin chains, particularly valuable for characterizing flexible conformations [14].
Cryo-Electron Microscopy (cryo-EM): High-resolution cryo-EM has visualized E3 ligases like TRIP12 in complex with ubiquitin chains, revealing mechanistic details of linkage-specific chain formation [96].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Studying Atypical Ubiquitin Linkages
| Reagent Category | Specific Examples | Research Applications | Key Features |
|---|---|---|---|
| Ubiquitin Mutants | K-to-R mutants (K6R, K11R, K27R, K29R, K33R, K48R, K63R) [97] | Linkage determination, linkage-specific function studies | Prevent chain formation via specific lysine |
| Ubiquitin Mutants | K-Only mutants (K6-only, K11-only, K27-only, K29-only, K33-only, K48-only, K63-only) [97] | Verification of linkage specificity, defined chain formation | Restrict chain formation to single lysine |
| E3 Ligase Tools | TRIP12 proteins and constructs [96] | K29-linked chain formation studies | Forms K29 linkages and K29/K48-branched chains |
| Inhibitors | ONX-0914 (β5i/LMP7 inhibitor) [98] | Immunoproteasome functional studies | Specific inhibition of immunoproteasome activity |
| Cell Line Models | Ubiquitin replacement cell lines [62] | Linkage-specific functional studies in cells | Conditional abrogation of specific linkage types |
Emerging Concepts and Future Perspectives
The study of atypical ubiquitin linkages continues to evolve with several emerging concepts reshaping our understanding:
Branched Ubiquitin Chains: Recent research has highlighted the importance of branched ubiquitin chains containing atypical linkages. For example, TRIP12 forms branched K29/K48-linked chains, while other E3s create various hybrid branched structures [96] [12]. These branched chains appear to represent specialized signals with unique functions beyond their linear counterparts.
Linkage Collaboration: There is growing evidence that E3 ligases collaborate to synthesize complex ubiquitin architectures. Pairs of E3s with distinct linkage specificities can work together to sequentially modify substrates, as demonstrated for Ufd4 and Ufd2 in yeast that collaborate to synthesize branched K29/K48 chains [12].
Chromatin Regulation: K29-linked ubiquitylation has been strongly associated with chromosome biology and has been identified as the essential degradation signal for the H3K9me3 methyltransferase SUV39H1, thereby regulating epigenome integrity [62]. This reveals a crucial connection between atypical ubiquitin linkages and epigenetic control.
The ongoing development of more specific reagents, including linkage-specific antibodies and chemical probes, will continue to accelerate our understanding of these complex signaling systems. Furthermore, the application of advanced structural techniques and single-molecule approaches will provide unprecedented insights into the dynamic nature of atypical ubiquitin linkages in cellular regulation.
Ubiquitylation is an essential post-translational modification that controls the stability, activity, and interaction properties of eukaryotic proteins. The complexity of ubiquitin signaling stems from the ability of ubiquitin to form diverse polymer chains, in which the C-terminus of one ubiquitin molecule is conjugated to one of the seven lysine residues (K6, K11, K27, K29, K33, K48, K63) or the N-terminal methionine (M1) of another ubiquitin molecule [12]. While early research focused on homogeneous chains, recent advances have revealed that branched ubiquitin chains—comprised of ubiquitin subunits simultaneously modified on at least two different acceptor sites—dramatically expand the complexity and functional scope of the ubiquitin code [12] [99]. These branched architectures are not simply structural curiosities but represent specialized signals that control critical cellular processes from cell cycle progression to immune signaling [100] [33]. This review examines the current understanding of branched ubiquitin chains involving the atypical linkages K6, K27, and K29, with particular focus on their assembly mechanisms, functional roles, and the experimental approaches enabling their study.
Architecture and Classification of Branched Ubiquitin Chains
Branched ubiquitin chains are classified by their linkage composition and architectural organization. Unlike homogenous chains composed of a single linkage type, or mixed chains containing multiple linkages but with each ubiquitin modified at only one site, branched chains contain at least one ubiquitin monomer modified simultaneously at two or more distinct sites [12]. The potential structural diversity is nearly limitless, as branch points can be initiated at distal, proximal, or internal ubiquitins within a chain, and the order of linkage synthesis can generate different architectures even with identical linkage compositions [12]. For instance, branched K11/K48 chains can be assembled by the APC/C through the addition of K11 linkages onto pre-existing K48-linked chains, whereas UBR5 creates the same linkage combination through the reciprocal order of addition [12].
Table 1: Documented Branched Ubiquitin Chains Involving Atypical Linkages
| Branch Linkage Combination | Reported Functions | Synthesizing Enzymes | Cellular Context |
|---|---|---|---|
| K6/K48 [12] | Unidentified functions | NleL (bacterial E3), Parkin [12] | In vitro and cellular systems |
| K6/K11 [12] | Unidentified functions | Not specified [12] | Detected in vitro or in cells |
| K27/K29 [12] | Unidentified functions | Not specified [12] | Detected in vitro or in cells |
| K29/K48 [12] | Protein degradation (UFD pathway) [12] | Ufd4 and Ufd2 collaboration (yeast) [12] | Ubiquitin Fusion Degradation pathway |
| K29/K33 [12] | Unidentified functions | Not specified [12] | Detected in vitro or in cells |
| K48/K63 [12] [28] | NF-κB signaling, apoptotic regulation [12] | TRAF6/HUWE1, ITCH/UBR5 collaboration [12] | Innate immune signaling, apoptosis |
The notation system for branched ubiquitin chains enables precise description of these complex polymers. In this system, ubiquitin units are connected by an en dash (−) with the distal-end ubiquitin placed to the left and the proximal ubiquitin (attached to the substrate) to the right [28]. Specific linkage residues are indicated as superscripts, and multiple ubiquitins branching from a single proximal ubiquitin are indicated with brackets. For example, a branched tri-Ub with two distal ubiquitins linked to K48 and K63 of a proximal ubiquitin is written as Ub[Ub]–⁴⁸,⁶³Ub or [Ub]₂–⁴⁸,⁶³Ub [28].
Branched Chains Involving K6, K27, and K29 Linkages
K6-Linked Branched Chains
K6-linked branched ubiquitin chains remain among the least characterized, though several synthetic enzymes have been identified. The bacterial HECT-like E3 ligase NleL can assemble branched K6/K48 chains in the presence of a single E2 enzyme [12]. Additionally, the RBR family E3 ligase Parkin, mutations in which cause early-onset Parkinson's disease, has been shown to synthesize branched K6/K48 chains [12]. This is consistent with previous reports that Parkin generates chains of complex topology including multiple linkage types [12]. While the precise physiological functions of K6-branched chains remain largely unexplored, their association with Parkin suggests potential roles in mitochondrial quality control and mitophagy, processes known to involve K6-linked ubiquitination in a proteolysis-independent manner [24].
K27-Linked Branched Chains
K27-linked ubiquitin chains have emerged as important regulators of innate immune signaling, and evidence suggests they can be incorporated into branched architectures. K27-linked chains function in balancing activation and inhibition of immune signaling pathways [33]. For instance, TRIM23 conjugates K27-linked chains to NEMO (NF-κB essential modulator), which is required for induction of NF-κB and IRF3 upon RIG-I-like receptor activation [33]. These K27-linked chains subsequently serve as platforms for recruiting additional regulatory factors. One mechanism involves Rhbdd3, which binds K27-linked chains on NEMO, leading to its own K27-linked ubiquitination and recruitment of the deubiquitinase A20, which then removes K63-linked chains from NEMO to prevent excessive NF-κB activation [33]. This intricate regulation demonstrates how branched chains containing K27 linkages might fine-tune immune responses.
K29-Linked Branched Chains
K29-linked ubiquitin chains participate in several documented branched structures with defined biological functions. In the ubiquitin fusion degradation (UFD) pathway in yeast, branched K29/K48 chains are synthesized through collaboration between Ufd4 and Ufd2 E3 ligases [12]. Ufd4 initially modifies substrates with K29-linked chains, which are then recognized by Ufd2 through two loops in its N-terminal domain, enabling Ufd2 to initiate branching by adding multiple K48-linked ubiquitins to the chain [12]. This modification targets substrates for proteasomal degradation. Additionally, RNF34 assembles branched chains containing both K27 and K29 linkages on MAVS (mitochondrial antiviral-signaling protein), inducing autophagy-mediated degradation of MAVS and thereby restricting the type I interferon response [8]. This identifies a role for K29-containing branched chains in negative regulation of antiviral signaling.
Synthesis Mechanisms and Enzymatic Machinery
The assembly of branched ubiquitin chains involves specialized enzymatic mechanisms that often require collaboration between different ubiquitylation enzymes. These mechanisms can be broadly categorized into multi-enzyme collaboration and single-E3 synthesis.
Collaborative Synthesis by E3 Pairs
A common theme in branched chain assembly is collaboration between pairs of E3 ligases with distinct linkage specificities [12]. The synthesis of branched K29/K48 chains by Ufd4 and Ufd2 in yeast represents a classic example of this mechanism, where one E3 recognizes and builds upon the chain type synthesized by its partner [12]. Similarly, during NF-κB signaling, TRAF6 and HUWE1 collaborate to produce branched K48/K63 chains [12]. In this case, HUWE1 attaches K48 linkages to unbranched K63-linked chains synthesized by TRAF6 by recognizing K63 linkages through its UIM and UBA domains [12]. Another collaborative pair, ITCH and UBR5, generates branched K48/K63 chains during apoptotic responses through a mechanism involving binding of K63-linked chains conjugated by ITCH to the UBA domain of UBR5, a K48-specific E3 [12]. This collaboration can efficiently convert a non-degradative K63-linked signal to a degradative K48-branched mark, providing a temporal regulatory mechanism for controlling signaling protein activation and inactivation.
Synthesis by Individual E3 Ligases
Some individual E3 ligases possess the intrinsic ability to synthesize branched chains, either by recruiting multiple E2 enzymes with different linkage specificities or through structural features that enable branching with a single E2. The anaphase-promoting complex/cyclosome (APC/C), a multisubunit RING E3, cooperates with two different E2s (UBE2C and UBE2S) to form branched K11/K48 chains on substrates during mitosis [12]. In this mechanism, UBE2C first attaches short chains containing mixed K11, K48, and K63 linkages to APC/C substrates, after which the K11-specific E2 UBE2S adds multiple K11 linkages to these short chains, resulting in branched K11/K48 polymers [12]. The APC/C engages UBE2C and UBE2S differently to create unique catalytic architectures that promote distinct stages of chain initiation and branching [12]. For individual E3s that assemble branched polymers with a single E2, the mechanisms are less clear but may involve intrinsic structural features. For instance, the HECT E3s WWP1 and UBE3C, both capable of forming branched chains, contain non-covalent ubiquitin-binding sites within or adjacent to their catalytic HECT domains that may facilitate chain branching [12].
Figure 1: Mechanisms for synthesis of branched ubiquitin chains. Collaborative synthesis often involves two E3 ligases working with different E2 enzymes, while single-E3 synthesis can occur through recruitment of multiple E2s or intrinsic branching capability with a single E2.
Functional Roles of Branched Ubiquitin Chains
Branched ubiquitin chains execute diverse cellular functions, often enhancing or modifying the outcomes mediated by homotypic chains. Key functional roles include:
Enhanced Proteasomal Degradation
Branched ubiquitin chains containing atypical linkages can significantly enhance substrate recognition and degradation by the proteasome. Research on the APC/C demonstrated that it efficiently synthesizes branched conjugates containing multiple blocks of K11-linked chains, which strongly enhance substrate recognition by the proteasome compared to homogenous chains [100]. These branched conjugates drive degradation of cell-cycle regulators during early mitosis, particularly under challenging conditions when degradation efficiency becomes critical [100]. This function appears conserved in yeast, where genetic analysis of K11R ubiquitin mutants revealed strong genetic interactions with a subunit of the APC, suggesting a role in cell cycle regulation [24]. Biochemical studies confirmed that yeast APC modifies substrates with K11-linkages in vitro, and these chains contribute to normal APC-substrate turnover in vivo [24].
Regulation of Innate Immune Signaling
Branched chains incorporating atypical linkages play crucial roles in regulating antiviral innate immune responses. Multiple E3 ligases assemble branched chains on immune signaling components to either activate or inhibit signaling pathways [8] [33]. For instance, RNF34 assembles branched chains containing K27 and K29 linkages on MAVS, inducing autophagy-mediated degradation of MAVS and thereby restricting the type I interferon response [8]. Similarly, TRIM23 conjugates K27-linked chains to NEMO, required for induction of NF-κB and IRF3 upon RLR activation [33]. The same E3 also auto-ubiquitinates with K27-linked chains, leading to TBK1 activation and induction of antiviral autophagy [33]. These examples illustrate how branched chains containing atypical linkages can both promote and inhibit immune signaling depending on the specific context.
Coordination of Complex Signaling Outcomes
The presence of multiple linkage types within branched ubiquitin chains enables the integration of different signals and coordination of complex cellular decisions. Branched chains can be edited by deubiquitinases (DUBs) that selectively remove specific linkages, thereby refining the ubiquitin code rather than terminating signaling entirely [99]. For example, UCH37, activated by binding to the proteasomal lid subunit RPN13, removes K48 linkages from branched ubiquitin molecules while leaving the variable chain intact [99]. This editing function can convert a degradative signal to a non-degradative one, adding another layer of regulation to branched chain signaling.
Table 2: Functional Roles of Atypical Linkages in Branched Ubiquitin Chains
| Linkage | Known Branch Partners | Primary Functional Contexts | Representative E3 Ligases |
|---|---|---|---|
| K6 | K48 [12] | Mitophagy, DNA damage response [12] [24] | Parkin, NleL, BRCA1-BARD1 [12] [24] |
| K27 | K29, K48, K63 [12] [8] | Innate immune signaling, inflammatory regulation [8] [33] | TRIM23, RNF34, MARCH8 [8] [33] |
| K29 | K27, K33, K48 [12] | Protein degradation (UFD), immune regulation, mRNA stability [12] [8] [24] | Ufd4, UBR5, RNF34 [12] [8] |
Methodologies for Studying Branched Ubiquitin Chains
Analytical and Proteomic Approaches
Advanced mass spectrometry techniques have been developed to identify and quantify branched ubiquitin chains. The Ub-AQUA-PRM (Ubiquitin Absolute Quantification by Parallel Reaction Monitoring) assay enables quantification of all ubiquitin chain types in high-throughput LC-MS/MS runs [21]. This approach has revealed tissue-specific enrichment of atypical ubiquitin chains; for example, K33-linked chains are enriched in contractile tissues like heart and muscle [21]. Middle-down mass spectrometry and "ubiquitin clipping" methods have also proven valuable for deciphering complex ubiquitin chain architectures, including branched conjugates [99]. These proteomic approaches can be complemented by linkage-specific antibodies, though their ability to distinguish branched from mixed chains is often limited.
Genetic Screening Approaches
Genetic interaction screening provides a powerful approach to identify physiological functions of specific ubiquitin linkages. Systematic genetic array (SGA) analysis in yeast combining gene deletions with lysine-to-arginine ubiquitin mutants has identified pathways regulated by specific chain types [24]. For example, K11R mutants showed strong genetic interactions with threonine biosynthetic genes, leading to the discovery that K11-linkages are important for threonine import [24]. The same screening approach also revealed a role for K11-linkages in APC-mediated cell cycle regulation in yeast, similar to their known function in vertebrates [24].
Figure 2: Methodological approaches for studying branched ubiquitin chains. Multiple complementary techniques are required to identify branched chains, characterize their functions, and elucidate their synthesis mechanisms.
Biochemical Reconstitution and Structural Biology
In vitro reconstitution of branched chain synthesis using purified E1, E2, and E3 enzymes has been instrumental for understanding the mechanisms of branch formation [12] [100]. Combined with structural techniques such as X-ray crystallography, NMR spectroscopy, and cryo-electron microscopy, these approaches have revealed how specific E3 ligases collaborate or intrinsically catalyze branching reactions [12] [28]. NMR studies of mixed-linkage chains containing both K48 and K63 linkages have demonstrated that each linkage within mixed chains retains the distinctive structural and recognition properties of its homotypic counterpart [28].
The Scientist's Toolkit: Key Research Reagents
Table 3: Essential Research Reagents for Studying Branched Ubiquitin Chains
| Reagent / Method | Function / Application | Key Features and Limitations |
|---|---|---|
| Linkage-specific ubiquitin mutants (K-to-R) [24] | Genetic analysis of linkage-specific functions | Enables study of physiological roles; may have compensatory effects |
| Ub-AQUA-PRM [21] | Absolute quantification of ubiquitin chain linkages | High-throughput capability; cannot distinguish branched from mixed chains |
| Linkage-specific antibodies [8] [33] | Detection of specific ubiquitin linkages | Variable specificity; limited ability to detect branching |
| Activity-based E3 profiling [99] | Identification of E3 ligases capable of synthesizing branched chains | Can reveal novel enzymatic activities; may not reflect cellular context |
| DUB linkage specificity profiling [99] | Characterization of branched chain disassembly | Identifies DUBs that edit rather than degrade branched chains |
| Bispecific antibodies [99] | Detection of specific branched chain architectures | Can potentially recognize unique epitopes formed by branching |
Branched ubiquitin chains containing K6, K27, and K29 linkages represent a sophisticated layer of regulation in the ubiquitin system. These complex polymers expand the signaling capacity of ubiquitination beyond what is possible with homotypic chains alone, enabling enhanced degradation signals, intricate immune regulation, and coordinated cellular decision-making. While significant progress has been made in identifying the enzymes that synthesize branched chains and some of their biological functions, major challenges remain. We still lack the ability to "sequence" ubiquitin chains as we do nucleic acids, limiting our understanding of the precise architecture and prevalence of branching in cells. Furthermore, the dynamics of branched chain assembly and disassembly in response to cellular signals remain poorly characterized. Future research developing new technologies to decipher the complete topology of ubiquitin chains and to manipulate specific branched chain types with temporal and spatial precision will greatly advance our understanding of these complex signals and their potential as therapeutic targets in human disease.
Atypical ubiquitin chains, characterized by linkages through lysine residues K6, K11, K27, K29, and K33, represent a sophisticated regulatory layer in cellular signaling that extends beyond the canonical K48- and K63-linked chains. Once considered poorly understood due to limited research tools, these atypical linkages are now recognized as critical players in pathogenic processes. This technical review synthesizes current evidence establishing how atypical ubiquitination drives molecular mechanisms in cancer progression, neurodegenerative pathology, and inflammatory dysregulation. We examine the specific E3 ligases and deubiquitinases (DUBs) that govern these processes, present methodological frameworks for their study, and discuss the emerging therapeutic potential of targeting atypical ubiquitin signaling. The compiled data underscore that decoding the ubiquitin code of atypical chains is essential for understanding disease etiology and developing novel intervention strategies.
Ubiquitination is a fundamental post-translational modification that regulates virtually all cellular processes in eukaryotes. The complexity of ubiquitin signaling stems from its ability to form diverse polymer chains through different linkage types between ubiquitin monomers. While canonical K48-linked chains primarily target substrates for proteasomal degradation and K63-linked chains regulate non-degradative signaling, the so-called "atypical" chains (linked via K6, K11, K27, K29, and K33) have emerged as specialized regulators with distinct functional consequences [8] [12].
The architectural diversity of ubiquitin chains extends beyond homotypic chains to include branched ubiquitin chains, where a single ubiquitin monomer is simultaneously modified on at least two different acceptor sites [12]. These complex structures create a sophisticated ubiquitin code that expands the biological information capacity of ubiquitin signaling. For instance, branched K11/K48 chains have been identified as particularly efficient degradation signals, while K29/K48 and K48/K63 branched chains play roles in regulating specific signaling pathways [12].
Understanding the functions of atypical ubiquitin chains has been challenging due to the historical lack of specific research tools. However, recent technological advances have revealed that these linkages are far from rare or insignificant—they constitute specialized regulatory systems that fine-tune critical cellular processes. When dysregulated, these systems contribute significantly to disease pathogenesis across multiple domains, including cancer, neurodegeneration, and inflammation [8] [101] [102].
Atypical Ubiquitination in Cancer Pathogenesis
The role of atypical ubiquitination in cancer manifests primarily through deregulation of transcription factors, tumor suppressors, and oncoproteins that control cell proliferation, apoptosis, and metastasis. The ubiquitin-proteasome system (UPS) has become an attractive target for cancer therapy, with several drugs already in clinical use and development [2].
Regulatory Mechanisms and Key Players
In cancer, atypical ubiquitin chains function as critical switches that control the stability and activity of key regulatory proteins. K11-linked chains have been implicated in cell cycle regulation, with UBE2C and UBE2S collaborating to form branched K11/K48 chains on substrates of the anaphase-promoting complex/cyclosome (APC/C) during mitosis [12]. Dysregulation of this process can lead to genomic instability and uncontrolled proliferation. The E3 ligase RNF26 utilizes K11-linked chains to stabilize STING, thereby modulating type I interferon production and potentially influencing tumor immunogenicity [8].
K27-linked ubiquitination has emerged as a particularly important regulator in cancer-related pathways. Multiple E3 ligases, including TRIM23, TRIM21, and AMFR, utilize K27 linkages to modulate innate immune signaling components such as NEMO, MAVS, and STING, creating a link between inflammatory signaling and cancer development [8]. The E3 ligase TRIM21 promotes K27-linked ubiquitination of MAVS, enhancing type I interferon production—a pathway with potential implications for cancer immunotherapy [8].
Table 1: Cancer-Relevant Atypical Ubiquitination Events
| Ubiquitin Linkage | E3 Ligase/DUB | Substrate | Functional Outcome in Cancer | References |
|---|---|---|---|---|
| K11-linked | RNF26 | STING | Inhibits STING degradation, increasing type I IFN production | [8] |
| K11/K48-branched | APC/C-UBE2C-UBE2S | Cell cycle regulators | Promotes mitotic progression; dysregulation causes genomic instability | [12] |
| K27-linked | TRIM21 | MAVS | Enhances type I IFN production; potential cancer immunology implications | [8] |
| K27-linked | AMFR | STING | Recruits TBK1, induces IRF3 activation and type I IFN production | [8] |
| K29-linked | SKP1-Cullin-Fbx21 | ASK1 | Induces IFNβ and IL-6 production; potential inflammation link | [8] |
Transcription Factor Regulation
Transcription factors represent critical control points in oncogenesis, and their regulation through atypical ubiquitination is increasingly recognized. In many cancers, tumor suppressors such as p53 undergo premature degradation due to abnormal ubiquitination, while oncogenic transcription factors may gain stability through the same mechanism [103]. The balance of ubiquitination and deubiquitination thus becomes a determining factor in tumor progression.
The E3 ligase MDM2 (HDM2 in humans) exemplifies this regulatory paradigm. While primarily known for its role in K48-linked ubiquitination of p53, emerging evidence suggests that MDM2 may also participate in atypical ubiquitination events that influence cancer development [2]. Additionally, ubiquitination can either enhance or inhibit transcription factor function depending on the cellular context, creating complex regulatory networks that differ between cancer types [103].
Atypical Ubiquitination in Neurodegenerative Disorders
Neurodegenerative diseases, including Parkinson's disease (PD) and Alzheimer's disease (AD), are characterized by the accumulation of misfolded protein aggregates and progressive neuronal loss. Atypical ubiquitin chains feature prominently in the pathology of these disorders, both as components of aggregates and as regulators of protein quality control mechanisms.
Protein Aggregation and Clearance
The presence of ubiquitin in pathological protein aggregates—such as Lewy bodies in PD, neurofibrillary tangles in AD, and inclusions in amyotrophic lateral sclerosis (ALS)—suggests a fundamental failure of protein homeostasis mechanisms [101]. Neurons, being post-mitotic cells, are particularly vulnerable to defects in protein clearance systems. Atypical ubiquitin chains contribute to both sides of this balance: regulating the degradation machinery and serving as signals on the accumulating proteins themselves.
K6-linked ubiquitination has been implicated in the cellular response to protein stress. The E3 ligase Parkin, mutations in which cause autosomal recessive juvenile PD, can synthesize branched K6/K48 chains, particularly in the context of mitophagy [12]. This suggests that K6 linkages may play a specialized role in mitochondrial quality control, a process critical for neuronal health. Similarly, K11-linked chains are associated with proteasomal degradation and may contribute to the clearance of misfolded proteins [101].
Table 2: Neurodegeneration-Associated Atypical Ubiquitination
| Ubiquitin Linkage | E3 Ligase/DUB | Substrate/Process | Role in Neurodegeneration | References |
|---|---|---|---|---|
| K6/K48-branched | Parkin | Mitophagy | Mutations cause early-onset Parkinson's disease | [12] |
| K27/K29 | RNF34 | MAVS degradation | Induces autophagy-mediated degradation; potential link to neuronal homeostasis | [8] |
| K33-linked | USP38 | TBK1 stabilization | Prevents TBK1 degradation, induces IRF3 activation; potential neuroinflammation role | [8] |
| Multiple linkages | UCH-L1 | α-synuclein dynamics | DUB activity linked to Parkinson's disease pathogenesis | [101] [104] |
| Multiple linkages | USP30 | PINK1/Parkin mitophagy | Negative regulator of mitophagy; overactivity linked to PD | [104] |
Mitochondrial Quality Control
The PINK1/Parkin pathway of mitophagy represents a particularly well-studied ubiquitin-dependent process in neurodegeneration. Upon mitochondrial damage, PINK1 stabilizes on the outer mitochondrial membrane and recruits Parkin, which then ubiquitinates numerous mitochondrial proteins [101]. While K63-linked chains have been extensively studied in this process, emerging evidence indicates that Parkin can synthesize multiple linkage types, including K6, K11, and K48, potentially forming branched chains that efficiently target mitochondria for degradation [12].
Dysregulation of this pathway directly contributes to neurodegeneration. Loss-of-function mutations in either PINK1 or Parkin cause autosomal recessive juvenile Parkinsonism, highlighting the importance of proper mitochondrial quality control in neuronal survival [101]. Additionally, several DUBs have been identified as regulators of this pathway, with USP30 emerging as a promising therapeutic target due to its ability to counter Parkin-mediated ubiquitination and dampen mitophagy [104].
Atypical Ubiquitination in Inflammatory and Immune Signaling
Inflammation represents a common thread connecting many chronic diseases, and atypical ubiquitin chains serve as crucial regulators of immune signaling pathways. The role of these linkages is particularly evident in the activation of pattern recognition receptors and subsequent induction of inflammatory cytokines and type I interferons.
Regulation of Innate Immune Signaling
The antiviral innate immune response provides a well-characterized system for understanding the functions of atypical ubiquitin chains. Linear (M1-linked) chains, assembled by the linear ubiquitin chain assembly complex (LUBAC), play a dual role in innate immunity: they potentiate NF-κB signaling while inhibiting type I interferon responses [8]. This is achieved through specific interactions with NF-κB essential modulator (NEMO), which contains a ubiquitin-binding domain with strong preference for linear chains [8].
K27-linked chains have emerged as particularly important regulators of antiviral signaling. Multiple E3 ligases, including TRIM23, TRIM26, and RNF185, utilize K27 linkages to modify key signaling components such as NEMO, cGAS, and STING, thereby modulating the production of type I interferons and proinflammatory cytokines [8]. The functional outcomes of K27-linked ubiquitination are context-dependent, with some modifications promoting signaling activation while others mediate inhibitory effects.
Table 3: Atypical Ubiquitination in Immune Signaling Pathways
| Ubiquitin Linkage | Regulatory Enzyme | Immune Pathway | Effect on Signaling | References |
|---|---|---|---|---|
| Linear/M1-linked | LUBAC | NF-κB signaling | Potentiates NF-κB activation through NEMO binding | [8] |
| K27-linked | TRIM23 | RIG-I/MDA-5 signaling | Activates IRF3 and NF-κB through NEMO modification | [8] [102] |
| K27-linked | RNF185 | cGAS-STING pathway | Induces IRF3 activation and type I IFN production | [8] |
| K27-linked | MARCH8 | MAVS | Induces autophagy-mediated degradation of MAVS, restricting IFN response | [8] |
| K29-linked | SKP1-Cullin-Fbx21 | ASK1 | Induces IFNβ and IL-6 production | [8] |
| K33-linked | USP38 | TBK1 | Prevents TBK1 degradation, enhances IRF3 activation | [8] |
Macrophage Polarization and Immunometabolism
Recent research has revealed that atypical ubiquitination plays a crucial role in macrophage polarization—the process by which macrophages adopt distinct functional phenotypes in response to environmental cues. The balance between pro-inflammatory M1-like and anti-inflammatory M2-like macrophages is tightly controlled by ubiquitin-mediated mechanisms [102].
The E3 ligase TRIM23 promotes NF-κB activation during viral infections by conjugating K27-linked chains to NEMO, thereby driving inflammatory M1-like polarization [102]. Conversely, the E3 ligase TRIM59 limits M1 polarization by ubiquitinating and degrading STAT1, while also maintaining M2 identity by restraining aberrant TNF-α production [102]. This intricate regulation demonstrates how atypical ubiquitin chains can integrate metabolic and inflammatory signals to shape immune responses.
The ubiquitin system also provides negative feedback mechanisms to prevent excessive inflammation. For instance, the DUBs A20 and CYLD remove activating ubiquitin chains from key adaptors in the NF-κB and MAPK pathways, thereby terminating signal transduction [102]. Dysregulation of these control mechanisms can lead to chronic inflammatory conditions and contribute to inflammatory pathology.
Experimental Approaches and Methodologies
Advancing our understanding of atypical ubiquitination requires specialized methodologies designed to detect, quantify, and functionally characterize these modifications. This section outlines key experimental protocols and research tools for studying atypical ubiquitin chains.
Detection and Characterization Methods
The identification of atypical ubiquitin linkages presents technical challenges due to their lower abundance compared to canonical chains and the limited specificity of available reagents. Current approaches include:
Linkage-Specific Antibodies: Development of antibodies that specifically recognize particular ubiquitin linkages has been instrumental in advancing the field. These reagents enable immunoblotting, immunohistochemistry, and immunoprecipitation of specific chain types. However, antibody availability remains limited for some atypical linkages, and validation of specificity is crucial.
Mass Spectrometry-Based Proteomics: Advanced proteomic techniques represent the gold standard for comprehensive ubiquitin linkage analysis. This typically involves expressing tagged ubiquitin in cells, enriching ubiquitinated proteins following proteasome inhibition, digesting with trypsin (which cleaves after arginine residues, leaving a di-glycine remnant on modified lysines), and analyzing by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Specialized data analysis tools are required to identify linkage types from spectral data.
Activity-Based Probes: Chemical probes that mimic ubiquitin or contain reactive groups can be used to profile the activity of specific E2 enzymes or E3 ligases involved in atypical chain formation. These tools are particularly valuable for studying the enzymatic machinery of atypical ubiquitination.
Functional Validation Approaches
Once identified, the functional significance of atypical ubiquitination events requires validation through multiple approaches:
Gene Editing: CRISPR/Cas9-mediated knockout of specific E3 ligases or DUBs suspected of regulating atypical ubiquitination provides a powerful approach to establish physiological function. Rescue experiments with wild-type and catalytically dead mutants can confirm specificity.
Linkage-Specific Mutants: Expression of ubiquitin mutants in which specific lysine residues are changed to arginine (preventing chain formation through that residue) can help establish the functional consequences of particular linkage types. However, interpretation can be complicated by potential redundancy and compensation mechanisms.
In Vitro Reconstitution: Purified E1, E2, and E3 enzymes can be combined with ubiquitin and candidate substrates in cell-free systems to directly demonstrate the formation of specific ubiquitin linkages and characterize the biochemical properties of the reaction.
Diagram 1: Experimental workflow for atypical ubiquitin chain characterization
Research Reagent Solutions
The study of atypical ubiquitination requires specialized reagents and tools. The following table summarizes key resources for investigating these post-translational modifications.
Table 4: Essential Research Reagents for Studying Atypical Ubiquitination
| Reagent Category | Specific Examples | Research Application | Considerations and Limitations |
|---|---|---|---|
| Linkage-specific antibodies | Anti-K27-linkage, Anti-K29-linkage, Anti-K11-linkage | Immunoblotting, immunofluorescence, immunoprecipitation | Variable commercial availability; require rigorous validation for specificity |
| Ubiquitin mutants | K6R, K11R, K27R, K29R, K33R | Functional studies to determine linkage-specific effects | Potential compensatory mechanisms may complicate interpretation |
| Activity-based probes | Ubiquitin-based probes with reactive groups | Profiling E1/E2/E3 activity and specificity | Require specialized chemistry and validation |
| Recombinant enzymes | E1, E2s (UBE2C, UBE2S), E3s (TRIM, RNF families) | In vitro ubiquitination assays | Proper folding and post-translational modifications may affect activity |
| DUB inhibitors | USP30, USP15 inhibitors | Functional studies of deubiquitination | Selectivity profiling essential to avoid off-target effects |
| Mass spectrometry standards | Heavy isotope-labeled ubiquitin | Quantitative proteomics of ubiquitination | Specialized instrumentation and expertise required |
The expanding landscape of atypical ubiquitination research reveals an intricate regulatory system with profound implications for human disease. Once overlooked, linkages such as K6, K11, K27, K29, and K33 are now recognized as specialized signals that fine-tune cellular processes in ways distinct from canonical ubiquitin chains. Their dysregulation contributes to pathogenesis across cancer, neurodegeneration, and inflammatory disorders through multiple mechanisms, including altered protein degradation, disrupted organelle quality control, and dysregulated immune signaling.
Therapeutic targeting of atypical ubiquitination pathways represents a promising frontier in drug development. Several strategies are emerging: inhibiting specific E3 ligases that drive pathological ubiquitination, developing DUB inhibitors to enhance the degradation of toxic proteins, and exploiting the ubiquitin system for targeted protein degradation using PROTAC (Proteolysis-Targeting Chimeras) technology [103] [104]. The clinical success of proteasome inhibitors in treating multiple myeloma and mantle cell lymphoma validates the UPS as a therapeutic target, paving the way for more precise interventions targeting specific components of the ubiquitin system.
Future research directions should focus on developing more specific tools to manipulate and monitor atypical ubiquitination events, elucidating the structural basis for linkage-specific recognition by ubiquitin-binding domains, and understanding the complex interplay between different ubiquitin linkage types in integrated physiological systems. As our methodological capabilities advance, so too will our understanding of how these sophisticated post-translational modifications coordinate cellular function in health and disease.
Conclusion
The study of K6, K27, and K29 ubiquitin linkages has moved from obscurity to the forefront of ubiquitin biology, revealing their essential and non-redundant roles in critical cellular pathways. These atypical chains are not merely minor variants but constitute a sophisticated regulatory layer within the ubiquitin code, with K27 emerging as a pivotal immune regulator, K6 functioning in organelle quality control, and K29 influencing signaling and stability processes. Future research must focus on developing more sensitive tools to detect these often-low-abundance modifications in physiological contexts, unraveling the complex mechanisms of branched chain formation involving these linkages, and identifying the full repertoire of enzymes that write, read, and erase these signals. The significant roles these chains play in immunity and cellular homeostasis present compelling opportunities for therapeutic intervention, particularly through the development of linkage-specific inhibitors that could offer precise control over pathological signaling pathways with minimal off-target effects.
References
- 1. Ubiquitin [https://en.wikipedia.org/wiki/Ubiquitin]
- 2. Progress in Anticancer Drug Development Targeting ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9737479/]
- 3. Beyond K48 and K63: non-canonical protein ubiquitination [https://cmbl.biomedcentral.com/articles/10.1186/s11658-020-00245-6]
- 4. The mechanism of linear ubiquitination in regulating cell ... [https://www.nature.com/articles/s41419-023-06183-3]
- 5. Emerging functions of branched ubiquitin chains - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7835216/]
- 6. 亲和分离在蛋白质泛素化修饰研究中的应用进展- PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9274849/]
- 7. Unraveling chain specific ubiquitination in cells using tandem ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12217038/]
- 8. The Role of Atypical Ubiquitin Chains in the Regulation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6987411/]
- 9. K29-Linked Ubiquitination of Transcription Regulators ... [https://pubmed.ncbi.nlm.nih.gov/40884253/]
- 10. Article K29-Selective Ubiquitin Binding Domain Reveals ... [https://www.sciencedirect.com/science/article/pii/S1097276515000908]
- 11. Ubiquitin Linkage-Specific Affimers Reveal Insights into K - 6 ... Linked [https://pmc.ncbi.nlm.nih.gov/articles/PMC5640506/]
- 12. Emerging functions of branched ubiquitin chains [https://www.nature.com/articles/s41421-020-00237-y]
- 13. USP8 regulates mitophagy by removing K6-linked ubiquitin ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4283406/]
- 14. Linkage via K27 bestows ubiquitin chains with unique ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4787624/]
- 15. The role of ubiquitination in tumorigenesis and targeted ... [https://www.nature.com/articles/s41392-020-0107-0]
- 16. Linkage via K27 Bestows Ubiquitin Chains with Unique ... [https://www.sciencedirect.com/science/article/pii/S0969212616000174]
- 17. Regulation of antiviral innate immune signaling and viral ... [https://www.nature.com/articles/s12276-021-00691-y]
- 18. Regulation of Cellular Antiviral Signaling by Modifications ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5833123/]
- 19. Article Quantitative Proteomics Reveals the Function of ... [https://www.sciencedirect.com/science/article/pii/S0092867409000890]
- 20. K29-Linked Ubiquitin Signaling Regulates Proteotoxic Stress ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8717942/]
- 21. Targeted proteomic analysis reveals enrichment of atypical ... [https://www.sciencedirect.com/science/article/pii/S1874391920303316]
- 22. Smurf1-Mediated Lys29-Linked Nonproteolytic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3811687/]
- 23. K29‐Linked Ubiquitination of Transcription Regulators ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12622412/]
- 24. Genetic analysis reveals functions of atypical polyubiquitin ... [https://elifesciences.org/articles/42955]
- 25. K29-linked free polyubiquitin chains affect ribosome ... [https://www.sciencedirect.com/science/article/pii/S1097276524004416]
- 26. Current methodologies in protein ubiquitination ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9373315/]
- 27. Review Ubiquitin—A structural perspective [https://www.sciencedirect.com/science/article/abs/pii/S1097276524010104]
- 28. Mixed-linkage ubiquitin chains send mixed messages - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3654000/]
- 29. The Length of a Ubiquitin Chain: A General Factor for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7384046/]
- 30. Unraveling chain specific ubiquitination in cells using ... [https://www.nature.com/articles/s41598-025-07242-9]
- 31. Ubiquitin Signaling: Extreme Conservation as a Source of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4197634/]
- 32. A widely distributed family of eukaryotic and bacterial ... [https://www.nature.com/articles/s41467-022-35244-y]
- 33. The Role of Atypical Ubiquitin Chains in the Regulation ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2019.00392/full]
- 34. TRIP12 structures reveal HECT E3 formation of K29 linkages ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12440805/]
- 35. Mechanism of Ubiquitin-Chain Formation by the human ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2696189/]
- 36. Recent progress in dissecting ubiquitin signals with ... [https://www.sciencedirect.com/science/article/abs/pii/S1367593122000722]
- 37. The Chemical Biology of Ubiquitin - PMC - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC10038182/]
- 38. Emerging tools and methods to study cell signalling ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12224918/]
- 39. Chemical approaches to explore ubiquitin-like proteins [https://pubs.rsc.org/en/content/articlehtml/2025/cb/d4cb00220b]
- 40. Super Enhanced Purification of Denatured-Refolded ... [https://www.sciencedirect.com/science/article/pii/S1535947624001427]
- 41. Use of a High-Affinity Ubiquitin-Binding Domain to Detect ... [https://pubmed.ncbi.nlm.nih.gov/40948893/]
- 42. Antibody toolkit reveals N-terminally ubiquitinated ... [https://www.nature.com/articles/s41467-021-24669-6]
- 43. Time-resolved in vivo ubiquitinome profiling by DIA-MS ... [https://www.nature.com/articles/s41467-021-25454-1]
- 44. Computationally Driven Top-Down Mass Spectrometry of ... [https://pubmed.ncbi.nlm.nih.gov/40766653/]
- 45. Localized K63 Ubiquitin Signaling Is Regulated by VCP/ ... [https://www.sciencedirect.com/science/article/pii/S1535947625000180]
- 46. Unbiased mapping of cereblon neosubstrate landscape by ... [https://www.nature.com/articles/s41467-025-62829-0]
- 47. Ubiquitin modifications | Cell Research [https://www.nature.com/articles/cr201639]
- 48. A genetic approach to study polyubiquitination in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6499080/]
- 49. A genetic approach to study polyubiquitination in ... [https://www.sciencedirect.com/science/article/abs/pii/S0076687918305160]
- 50. UFMylation System: Biological Functions, Molecular ... [https://pubmed.ncbi.nlm.nih.gov/41079645/]
- 51. Current methodologies in protein ubiquitination characterization [https://cellandbioscience.biomedcentral.com/articles/10.1186/s13578-022-00870-y]
- 52. Unraveling Chain specific Ubiquitination in Cells Using ... [https://lifesensors.com/unraveling-chain-specific-ubiquitination-in-cells-using-tandem-ubiquitin-binding-entities-in-a-high-throughput-assay/]
- 53. The dynamic duo: Combining NMR and small angle ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4093944/]
- 54. Fast ultra-selective 1H-15N 1D NMR spectroscopy unlocks ... [https://chemrxiv.org/engage/chemrxiv/article-details/6890a86923be8e43d6b583a0]
- 55. Elucidation of the Protein Folding Landscape by NMR [https://www.sciencedirect.com/science/article/abs/pii/S0076687905940111]
- 56. 19F NMR study of proteins with parallel incorporation ... [https://www.sciencedirect.com/science/article/pii/S277251622500018X]
- 57. Recent advancements in neutron scattering techniques for ... [https://www.sciencedirect.com/science/article/pii/S0079670025001200?dgcid=rss_sd_all]
- 58. Accelerated small angle neutron scattering algorithms for ... [https://pubs.rsc.org/en/content/articlehtml/2025/sm/d4sm01350f]
- 59. Deubiquitinase dynamics: methodologies for understanding ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12123204/]
- 60. Discovery and mechanism of K63-linkage-directed ... [https://www.nature.com/articles/s41589-024-01777-0]
- 61. Thioether-mediated protein ubiquitination in constructing affinity [https://pubmed.ncbi.nlm.nih.gov/40281337/]
- 62. Functional landscape of ubiquitin linkages couples K29- ... [https://link.springer.com/article/10.1038/s44318-025-00599-7]
- 63. Low-Abundance Protein Enrichment for Medical Applications [https://pmc.ncbi.nlm.nih.gov/articles/PMC10299117/]
- 64. Validated assays for the quantification of C9orf72 human ... [https://www.nature.com/articles/s41598-023-50667-3]
- 65. Editorial: Advanced Strategies for the Recognition, ... [https://www.frontiersin.org/journals/bioengineering-and-biotechnology/articles/10.3389/fbioe.2022.957878/full]
- 66. Structure and Function of HECT E3 Ubiquitin Ligases and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7500122/]
- 67. Site-directed multivalent conjugation of antibodies to ... [https://www.nature.com/articles/s41551-024-01342-z]
- 68. A General Guide for the Optimization of Enzyme Assay ... [https://www.sciencedirect.com/science/article/pii/S247255522206779X]
- 69. A General Guide for the Optimization of Enzyme Assay ... [https://pubmed.ncbi.nlm.nih.gov/30802413/]
- 70. The Proteasome Distinguishes between Heterotypic and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4533228/]
- 71. Assembly and Function of Heterotypic Ubiquitin Chains in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5669814/]
- 72. Ubiquitin chain conformation regulates recognition and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3605796/]
- 73. UbiREAD deciphers proteasomal degradation code of ... [https://www.sciencedirect.com/science/article/pii/S1097276525001522]
- 74. K27‐linked ubiquitylation promotes p97 substrate processing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9058539/]
- 75. K27-linked RORγt ubiquitination by Nedd4 potentiates Th17 ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11841259/]
- 76. Expression and Regulation of Deubiquitinase-Resistant ... [https://www.nature.com/articles/s41598-018-26364-x]
- 77. Structural basis for specific recognition of K6-linked ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8391078/]
- 78. SI200: Panel Customized Ubiquitin Chain Kit (K6, K11, K27 ... [https://lifesensors.com/product/si200-panel-customized-ubiquitin-chain-kit-k6-k11-k27-k29-k33-k48-k63-and-linear/]
- 79. VME probe Archieven - UbiQ [https://ubiqbio.com/product-tag/vme-probe/]
- 80. Atypical ubiquitin chains: new molecular signals. 'Protein ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2427391/]
- 81. Functional landscape of ubiquitin linkages couples K29- ... [https://pubmed.ncbi.nlm.nih.gov/41125851/?utm_source=FeedFetcher&utm_medium=rss&utm_campaign=None&utm_content=0eVY6Aiv8oxUcUuXiandtUKo6DYmALAxhjy0GtsF5VE&fc=None&ff=20251029211250&v=2.18.0.post22+67771e2]
- 82. Atypical K6-ubiquitin chains mobilize p97/VCP and the ... [https://www.sciencedirect.com/science/article/pii/S1097276523009541]
- 83. Structural visualization of HECT-type E3 ligase Ufd4 ... [https://www.nature.com/articles/s41467-025-59569-6]
- 84. Non-catalytic UBL2 domain directs deubiquitinase USP11 ... [https://www.sciencedirect.com/science/article/pii/S0021925825027760]
- 85. A framework for modeling human monogenic diseases by ... [https://research.arcadiascience.com/pub/resource-modeling-human-monogenic-diseases/release/3/]
- 86. CRISPR-based functional genomics tools in vertebrate ... [https://www.nature.com/articles/s12276-025-01514-0]
- 87. The Role of Atypical Ubiquitin Chains in the Regulation ... [https://pubmed.ncbi.nlm.nih.gov/32039206/]
- 88. The K48-K63 Branched Ubiquitin Chain Regulates NF-κB ... [https://www.sciencedirect.com/science/article/pii/S1097276516305639]
- 89. Structural basis of K11/K48-branched ubiquitin chain ... [https://www.nature.com/articles/s41467-025-64719-x]
- 90. and deubiquitination in cancer: from mechanisms to... Ubiquitination [https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-024-02046-3]
- 91. What are Atypical Ubiquitin Chains? [https://www.news-medical.net/life-sciences/What-are-Atypical-Ubiquitin-Chains.aspx]
- 92. Crosstalk between Ubiquitination and Other Post-translational ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7748009/]
- 93. Article Structural basis for specific recognition of K6-linked ... [https://www.sciencedirect.com/science/article/pii/S000634952100549X]
- 94. Regulation of proteostasis and innate immunity via ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10857905/]
- 95. The various roles of ubiquitin in Wnt pathway regulation - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3047798/]
- 96. TRIP12 structures reveal HECT E3 formation of K29 ... [https://www.nature.com/articles/s41594-025-01561-1]
- 97. Protocol for Determining Ubiquitin Chain Linkage [https://www.rndsystems.com/resources/protocols/determine-ubiquitin-chain-linkage]
- 98. Immunoproteasomes control activation of innate immune ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2022.982786/full]
- 99. Review Assembly and function of branched ubiquitin chains [https://www.sciencedirect.com/science/article/abs/pii/S0968000422000883]
- 100. Enhanced Protein Degradation by Branched Ubiquitin ... [https://www.sciencedirect.com/science/article/pii/S0092867414004127]
- 101. Ubiquitin signalling in neurodegeneration - Nature [https://www.nature.com/articles/s41418-020-00706-7]
- 102. Ubiquitination in macrophage plasticity: from inflammatory to ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12507854/]
- 103. Ubiquitination of transcription factors in cancer: unveiling ... [https://pubmed.ncbi.nlm.nih.gov/40227962/]
- 104. Deubiquitinating enzymes in parkinson's disease [https://pmc.ncbi.nlm.nih.gov/articles/PMC12593799/]