A Comprehensive Guide to Preventing Deubiquitination During Cell Lysis: Strategies for DUB Inhibitor Implementation in Proteostasis Research
This article provides researchers, scientists, and drug development professionals with a comprehensive framework for preserving cellular ubiquitination states during protein extraction.
A Comprehensive Guide to Preventing Deubiquitination During Cell Lysis: Strategies for DUB Inhibitor Implementation in Proteostasis Research
Abstract
This article provides researchers, scientists, and drug development professionals with a comprehensive framework for preserving cellular ubiquitination states during protein extraction. We explore the fundamental biology of deubiquitinating enzymes (DUBs) and their disruptive potential during cell lysis, present methodological approaches for implementing DUB inhibitors in experimental workflows, troubleshoot common challenges in inhibitor selection and optimization, and establish validation protocols for assessing ubiquitin preservation efficacy. By integrating foundational principles with practical applications, this guide aims to enhance experimental reproducibility and data quality in ubiquitin-proteasome system research, supporting advancements in targeted protein degradation therapeutics and proteostasis investigation.
Understanding DUB Biology and the Critical Need for Inhibition During Cell Disruption
Technical Troubleshooting Guide: Preserving the Ubiquitinome during Cell Lysis
A primary challenge in ubiquitin research is maintaining the integrity of the ubiquitinome during cell lysis, as the natural activity of deubiquitinating enzymes (DUBs) can rapidly erase these post-translational modifications. The table below outlines common experimental issues and their solutions.
Table 1: Troubleshooting Common Issues in Ubiquitinome Analysis
| Problem | Potential Cause | Recommended Solution | Principle |
|---|---|---|---|
| Rapid loss of ubiquitin signal | DUB activity during lysis | Add DUB inhibitors (e.g., 10-50 µM PR-619 [1] [2]) and alkylating agents (e.g., 5-20 mM NEM [3] [4]) to lysis buffer. | Irreversibly blocks the catalytic cysteine of most DUBs [5] [6]. |
| High background in ubiquitin pulldowns | Non-specific protein binding or inefficient capture | Use Tandem Ubiquitin-Binding Entities (TUBEs) instead of single domains for purification [3]. | TUBEs have ~100-1000x higher affinity for poly-ubiquitin chains, enabling efficient capture under native conditions [3]. |
| Inconsistent DUB inhibition | Reversible DUB inhibitors being diluted or inactivated | Use covalent, irreversible DUB inhibitors like RA-9 [1] to ensure sustained inhibition. | Compound exposes a carbonyl group to a nucleophilic attack from the SH- group of the catalytic cysteine, forming a permanent bond [1]. |
| Altered mass signatures in MS | Iodoacetamide (IAA) forming protein adducts [3] | Use N-Ethylmaleimide (NEM) as an alternative cysteine alkylating agent [3]. | NEM modifies cysteine residues without creating adducts that mimic a double glycine signature, preventing misinterpretation of mass spectrometry data [3]. |
| Loss of specific ubiquitin chain types | Preferential cleavage of certain linkages by co-purifying DUBs | Include linkage-specific DUB inhibitors if available, and perform lysis in the presence of TUBEs [3]. | TUBEs protect poly-ubiquitin chains from both proteasomal degradation and deubiquitinating activity present in cell extracts [3]. |
Frequently Asked Questions (FAQs)
Q1: Why is it so critical to inhibit DUBs during cell lysis for ubiquitination studies?
Deubiquitinating enzymes (DUBs) are highly active cysteine proteases that constantly reverse ubiquitination. During cell lysis, the compartmentalization that may regulate their activity is lost. Without immediate and potent inhibition, DUBs will rapidly remove ubiquitin chains from your protein substrates, leading to a significant underestimation of ubiquitination levels and potentially erroneous conclusions [5] [3]. The fundamental goal is to "freeze" the ubiquitination state of the proteome as it existed in the living cell at the moment of lysis.
Q2: What are the key advantages of using TUBEs over traditional ubiquitin pulldown methods?
TUBEs (tandem-repeated ubiquitin-binding entities) offer several key advantages [3]:
- Superior Affinity: By linking four ubiquitin-associated (UBA) domains in tandem, TUBEs bind tetra-ubiquitin with an affinity 100 to 1,000 times greater than single UBA domains.
- Native Condition Purification: Their high affinity allows for the purification of ubiquitylated proteins from cell extracts under native conditions, without the need for harsh denaturants.
- Built-in Protection: TUBEs physically shield ubiquitin chains from the activity of DUBs and the proteasome, preserving the ubiquitinome during the purification process. They are more effective than using cysteine protease inhibitors like iodoacetamide or NEM alone [3].
Q3: Are there any risks associated with using pan-DUB inhibitors like PR-619 in my experiments?
Yes, while pan-DUB inhibitors are powerful tools, their non-specific nature can induce complex and sometimes unintended cellular phenotypes that complicate data interpretation. For instance, PR-619 has been shown to inhibit cell adhesion and proliferation in lung cancer and mesothelioma cell lines. However, its effect on cell motility was cell line-specific, increasing motility in one line while decreasing it in another [1]. Furthermore, broad DUB inhibition induces ER stress, apoptosis, and autophagy due to the accumulation of ubiquitylated proteins [2]. Therefore, for functional studies, more specific DUB inhibitors are recommended once a target of interest is identified.
Q4: How does oxidative stress impact DUB activity, and how should this be controlled?
Reactive oxygen species (ROS) like H~2~O~2~ can reversibly inactivate many DUBs of the USP and UCH subfamilies by oxidizing the catalytic cysteine residue [5]. This is a regulatory mechanism in cells. To control for this in experiments, maintain consistent reducing conditions. The inclusion of reducing agents like DTT (dithiothreitol) in lysis buffers can reverse this oxidation and restore DUB activity [5]. If studying redox regulation of DUBs, omit DTT and carefully control the oxidative environment.
Essential Protocols
Protocol for Cell Lysis with Optimized DUB Inhibition
This protocol is designed to maximally preserve ubiquitin conjugates for downstream analysis like western blotting or mass spectrometry.
Materials:
- Lysis Buffer (e.g., RIPA or NP-40 based)
- Protease Inhibitor Cocktail (without DUB inhibitors)
- Phosphatase Inhibitor Cocktail (if needed)
- N-Ethylmaleimide (NEM), 1M stock in ethanol
- PR-619, 50 mM stock in DMSO
- TUBEs (commercially available with GST or other tags [3])
Procedure:
- Prepare Inhibitor-Enriched Lysis Buffer: Add NEM to a final concentration of 5-20 mM and PR-619 to a final concentration of 10-50 µM to the chilled lysis buffer immediately before use [3] [1] [2].
- Lyse Cells: Aspirate media from culture dishes and immediately add cold lysis buffer. Scrape cells and transfer the lysate to a microcentrifuge tube.
- Incubate: Maintain lysates on ice for 15-30 minutes with occasional vortexing.
- Clarify: Centrifuge at >12,000 x g for 15 minutes at 4°C to remove insoluble debris.
- Proceed to Analysis: The clarified lysate is now ready for protein quantification, immunoprecipitation, or direct analysis by western blot. For ubiquitin pulldowns, add TUBEs directly to the lysate.
Protocol for qHTS of DUB Inhibitors Using a Cell Lysate-Based AlphaLISA Assay
This protocol enables high-throughput screening for DUB inhibitors in a more physiologically relevant cell lysate environment [4].
Materials:
- HEK293 cells expressing HA-tagged DUB of interest
- Biotinylated Ubiquitin Vinyl Methyl Ester (biotin-UbVMe) probe [4]
- AlphaLISA Anti-HA Acceptor Beads
- AlphaLISA Streptavidin Donor Beads
- White 1536-well microplates
- Test compound library
Workflow: The assay relies on the covalent binding of the biotin-UbVMe probe to the active site of the HA-tagged DUB. This proximity brings the donor and acceptor beads together, generating a signal inhibited by active compounds.
Diagram 1: AlphaLISA DUB Assay Workflow
Procedure [4]:
- Prepare Lysate: Lyse cells expressing the HA-tagged DUB in a suitable buffer (potentially without DUB inhibitors to preserve activity).
- Dispense: Transfer lysate and test compounds to a 1536-well plate.
- Probe Incubation: Add the biotin-UbVMe probe and incubate to allow covalent labeling of the DUB.
- Bead Addition: First, add Anti-HA Acceptor Beads, followed by Streptavidin Donor Beads.
- Read Plate: Illuminate the plate at 680nm and measure light emission at 615nm. A reduction in signal compared to a DMSO control indicates inhibition of the DUB's activity.
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Reagents for Ubiquitinome Preservation and Analysis
| Reagent | Function | Key Feature |
|---|---|---|
| PR-619 | Pan-DUB inhibitor, cell-permeable [1] [2]. | Broad-spectrum, inhibits many USPs, UCHs, and OTUs. Useful for initial proof-of-concept studies. |
| N-Ethylmaleimide (NEM) | Cysteine alkylating agent [5] [3]. | Irreversibly inactivates cysteine-dependent DUBs. Preferred over IAA for mass spectrometry. |
| TUBEs (Tandem Ubiquitin-Binding Entities) | High-affinity capture of poly-ubiquitinated proteins [3]. | Protects ubiquitin chains from DUBs and the proteasome during purification; enables native purifications. |
| Ubiquitin-Vinyl Methyl Ester (UbVMe) | Activity-based DUB probe [4]. | Covalently labels active site of DUBs; used for activity profiling and inhibitor screening (e.g., in AlphaLISA). |
| PYR-41 | Ubiquitin E1 Activating Enzyme inhibitor [2]. | Blocks the entire UPS upstream; useful as a control to confirm UPS-related phenotypes. |
| Biotin-UbVMe | Functionalized DUB probe for affinity applications [4]. | Contains a biotin tag for pulldown or bead-based assays (e.g., AlphaLISA) to detect active DUBs. |
Signaling Pathways in DUB Inhibition
Broad-spectrum DUB inhibition triggers a defined cellular stress response. The following diagram summarizes the key pathway activated upon treatment with inhibitors like PR-619, leading to cell death.
Diagram 2: Cellular Response to DUB Inhibition
Deubiquitinating enzymes (DUBs) constitute a critical regulatory protein superfamily that opposes the action of ubiquitin ligases by cleaving ubiquitin from protein substrates [7] [8]. This reversible process governs protein stability, localization, activity, and interactions, making DUBs essential regulators of cellular homeostasis [7] [9]. The human genome encodes approximately 100 DUBs, which are classified into two main classes based on their catalytic mechanisms: cysteine proteases and metalloproteases [7] [10]. These enzymes perform several essential functions, including processing ubiquitin precursors, removing degradative and non-degradative ubiquitin signals from target proteins, editing ubiquitin chain types, and maintaining the cellular pool of free ubiquitin [7] [8] [9]. Understanding these enzyme families is particularly crucial for experiments aimed at preserving ubiquitin signaling states during cell lysis, where uncontrolled deubiquitination can compromise experimental results.
DUB Classification and Biochemical Mechanisms
Cysteine Protease DUBs
Cysteine protease DUBs represent the majority of deubiquitinating enzymes and utilize a catalytic triad or dyad involving a cysteine residue for nucleophilic attack on the isopeptide bond [7] [9]. This class encompasses several families with distinct structural and functional characteristics, as detailed in Table 1.
Table 1: Major Cysteine Protease DUB Families
| Family | Representative Members | Catalytic Mechanism | Key Characteristics | Substrate/Linkage Preferences |
|---|---|---|---|---|
| USP | USP1, USP7, USP10, USP28 | Catalytic triad (Cys, His, Asp/Asn) [7] | Largest and most diverse family; often regulated by protein-protein interactions and domains [7] [11] | Varied; often specific to particular substrates or chain types [10] |
| UCH | UCH-L1, UCH-L3, UCH-L5 | Catalytic triad (Cys, His, Asp/Asn) [7] | Small molecules with narrow active site clefts; prefer small protein adducts [8] | Primarily cleaves ubiquitin from small nucleophiles and peptide substrates [7] |
| OTU | OTUB1, OTUD5, A20, OTULIN | Catalytic triad (Cys, His, Asp/Asn) [7] | Often exhibit high linkage specificity; regulated by oxidative stress and protein interactions [9] [11] | Specific for particular ubiquitin chain types (e.g., K48, K11, linear) [11] |
| MJD | Ataxin-3, Ataxin-3L | Catalytic triad (Cys, His, Asp/Asn) [7] | Josephin domain proteins; some associated with neurological disorders [7] [8] | Prefer K63-linked chains (Ataxin-3) [8] |
| MINDY | N/A | Catalytic triad (Cys, His, Asp/Asn) [12] | Recently identified family [12] | Prefer K48-linked ubiquitin chains [13] |
| ZUFSP | N/A | Catalytic triad (Cys, His, Asp/Asn) [13] | Recently identified family [13] | Specific for K63-linked polyubiquitin [13] |
Metalloprotease DUBs
The metalloprotease DUBs represent a distinct mechanistic class with a single family:
- JAMM/MPN+ Proteases: These zinc-dependent metalloproteases constitute the only metalloprotease class among DUBs [7] [9]. Unlike cysteine proteases, they employ a catalytic mechanism that coordinates zinc ions with histidine, aspartate, and serine residues to activate water molecules for nucleophilic attack on the isopeptide bond [7]. Representative members include POH1 (yeast homologue Rpn11), AMSH, and AMSH-LP [8] [10]. These enzymes often require integration into macromolecular complexes for full activity, such as the proteasome (POH1/Rpn11) or endosomal sorting complexes (AMSH) [10] [11]. Their metal-dependent mechanism renders them insensitive to cysteine-directed inhibitors, a crucial consideration for experimental design.
DUB Domain Architecture and Regulation
DUBs frequently contain accessory domains beyond their catalytic domains that regulate their activity, specificity, and subcellular localization [7]. Key regulatory domains include:
- Ubiquitin-Like (UBL) Domains: Present in many USPs, these domains can autoinhibit catalytic activity or facilitate proteasomal localization [7] [11].
- DUSP Domains: Found in several USPs, these tripod-like domains may contribute to substrate recognition and protein-protein interactions [7].
- Zinc Finger Domains: Various zinc finger motifs (e.g., ZnF-UBP, ZnF-MYND) mediate specific ubiquitin binding and substrate recognition [7].
DUB activity is tightly regulated through multiple mechanisms, including post-translational modifications, subcellular localization, protein-protein interactions, and oxidative inactivation [9] [11]. For instance, the catalytic activity of USP7 is enhanced through interactions with its C-terminal UBL domains and binding partners like GMP synthase [11]. Similarly, OTULIN specificity for linear ubiquitin chains is governed by unique interactions with the N-terminal methionine of ubiquitin [11]. Understanding these regulatory mechanisms is essential for designing effective strategies to control DUB activity during experimental procedures.
The Scientist's Toolkit: Research Reagent Solutions
Selecting appropriate reagents is fundamental for successful DUB research, particularly for inhibiting DUB activity during cell lysis. Table 2 summarizes key reagents and their applications.
Table 2: Essential Research Reagents for DUB Inhibition and Analysis
| Reagent Category | Specific Examples | Mechanism of Action | Primary Applications | Important Considerations |
|---|---|---|---|---|
| Broad-Spectrum Cysteine DUB Inhibitors | PR-619 [14] [15] | Inhibits cysteine proteases but not metalloproteases [14] | Cell lysis preparation; global DUB inhibition studies | Batch-to-batch variability in activity reported [14]; not suitable for JAMM metalloproteases |
| Ubiquitin-Activating Enzyme (E1) Inhibitors | TAK243 [14] | Blocks ubiquitin activation, preventing all ubiquitination | Control for ubiquitination dynamics; depletes ubiquitin conjugates | Affects entire ubiquitin system; not specific to DUBs |
| Activity-Based Probes | Biotin-UbVMe [12], Biotin-Ub-PA [15] | Covalently labels active DUBs with specified warheads | DUB profiling, identification, and validation in cell lysates | Confirms DUB activity status; useful for competitive assays |
| Linkage-Specific Antibodies | K48- and K63-linkage antibodies [14] | Immunodetection of specific ubiquitin chain types | Monitoring specific ubiquitin signals by immunoblotting | Verify specificity; some cross-reactivity may occur |
| Proteasome Inhibitors | MG132, Bortezomib, Carfilzomib [14] | Inhibit proteasomal degradation of ubiquitinated proteins | Stabilizing ubiquitinated substrates; studying degradation-independent ubiquitination | Does not prevent deubiquitination by DUBs |
Experimental Protocols for DUB Inhibition During Cell Lysis
Comprehensive DUB Inhibition Protocol for Cell Lysis
Principle: This protocol ensures maximal preservation of ubiquitin conjugates during cell lysis by combining broad-spectrum cysteine DUB inhibitors with appropriate buffer conditions.
Reagents:
- Lysis Buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 1 mM EDTA
- Protease Inhibitor Cocktail (without DUB inhibitors)
- Phosphatase Inhibitors (if studying phospho-ubiquitin)
- 10× DUB Inhibitor Cocktail: 10 mM PR-619 in DMSO (or equivalent concentration)
- N-Ethylmaleimide (NEM): 500 mM stock in ethanol (optional, for additional cysteine protection)
- Dithiothreitol (DTT): 1 M stock (AVOID in lysis buffer as it inactivates cysteine-directed inhibitors)
Procedure:
- Prepare complete lysis buffer freshly by adding protease inhibitors and 1× final concentration of DUB inhibitor cocktail (e.g., 1:1000 dilution of 10 mM PR-619 stock for 10 µM final concentration).
- Pre-chill complete lysis buffer on ice.
- Harvest cells by rapid centrifugation (500 × g, 3 min, 4°C) and wash once with cold PBS.
- Lyse cell pellet in appropriate volume of complete lysis buffer (typically 3-5× pellet volume).
- Incubate on ice for 15-30 minutes with occasional vortexing.
- Clarify lysate by centrifugation (14,000 × g, 15 min, 4°C).
- Transfer supernatant to fresh pre-chilled tube and proceed immediately to downstream applications.
- For long-term storage, flash-freeze aliquots in liquid nitrogen and store at -80°C.
Critical Considerations:
- DUB inhibitors must be present BEFORE cell disruption to prevent immediate deubiquitination.
- Avoid reducing agents (DTT, β-mercaptoethanol) in lysis buffers as they will inactivate cysteine-directed inhibitors.
- Include appropriate controls: DMSO-only treated lysates to demonstrate endogenous DUB activity.
- For metalloprotease JAMM family DUBs, additional specific inhibitors (e.g., orthophenanthroline) may be required as they are insensitive to cysteine protease inhibitors.
Validation Protocol: Assessing DUB Inhibition Efficiency
Principle: Verify the effectiveness of DUB inhibition by monitoring ubiquitin conjugate accumulation through immunoblotting.
Procedure:
- Prepare parallel cell lysates with and without DUB inhibitor cocktail.
- Separate equal protein amounts (20-30 µg) by SDS-PAGE (4-12% gradient gels recommended).
- Transfer to PVDF membrane and immunoblot with specified antibodies.
- Probe with anti-ubiquitin antibody to visualize global ubiquitin conjugate accumulation.
- Reprobe with anti-β-actin or other loading control antibodies for normalization.
- Compare ubiquitin signal intensity between inhibited and non-inhibited samples.
Expected Results: Successful DUB inhibition should yield significantly enhanced high-molecular-weight ubiquitin smears in inhibitor-treated samples compared to controls [14].
Troubleshooting:
- Weak ubiquitin signal: Increase inhibitor concentration; verify inhibitor solubility and activity.
- Excessive background: Optimize antibody concentrations; include no-primary-antibody controls.
- Incomplete inhibition: Combine multiple inhibitor classes; ensure rapid lysis and inhibitor penetration.
DUB Inhibitor Mechanisms and Experimental Workflow
The following diagram illustrates the strategic approach to preserving ubiquitin conjugates during cell lysis through DUB inhibition, integrating key reagents and validation steps.
Frequently Asked Questions (FAQs)
Q1: Why is DUB inhibition critical during cell lysis, and what happens if inhibitors are omitted? A: DUBs are highly active and can rapidly remove ubiquitin modifications from substrates upon cell disruption. Without inhibition, this results in significant loss of ubiquitin signals, compromising downstream analyses like ubiquitin immunoblotting, proteomic studies, or activity assays. Research shows that DUBs can process the bulk of ubiquitin conjugates within 3 hours after inhibition of new ubiquitination [14].
Q2: What are the key differences between cysteine protease and metalloprotease DUB inhibitors? A: Cysteine protease inhibitors (e.g., PR-619) target the catalytic cysteine residue in USP, UCH, OTU, MJD, MINDY, and ZUFSP families through covalent or non-covalent mechanisms. Metalloprotease inhibitors (e.g., 1,10-phenanthroline) chelate zinc ions essential for JAMM family enzyme activity. A comprehensive inhibition strategy requires both approaches since neither inhibitor class affects the other DUB family [7] [14].
Q3: How do I select the appropriate DUB inhibitor concentration for my experiment? A: Optimal concentration depends on cell type, abundance of target DUBs, and experimental goals. For broad-spectrum inhibition during lysis, start with 10-50 µM for PR-619 [14] [15]. Perform dose-response experiments using ubiquitin immunoblotting to visualize ubiquitin conjugate accumulation. Include DMSO-only controls to assess inhibition efficiency.
Q4: Can I use DTT or β-mercaptoethanol in lysis buffers with cysteine DUB inhibitors? A: No. Reducing agents react with and inactivate cysteine-directed inhibitors. Use alternative protease inhibitor cocktails without reducing agents. If protein reduction is essential, consider adding inhibitors after reduction or using non-covalent inhibitors unaffected by reducing conditions.
Q5: How can I validate that my DUB inhibition strategy is working effectively? A: Employ multiple validation approaches:
- Monitor high-molecular-weight ubiquitin conjugates by immunoblotting [14]
- Use activity-based probes (e.g., biotin-UbVMe) to assess residual DUB activity in lysates [12] [15]
- Analyze specific ubiquitin chain types (K48, K63) with linkage-specific antibodies [14]
- Compare with E1 inhibitor (TAK243) treated samples to distinguish DUB-mediated effects [14]
Q6: Are there specific considerations for studying JAMM metalloprotease DUBs? A: Yes. JAMM family DUBs (e.g., POH1, AMSH) are insensitive to cysteine protease inhibitors. For comprehensive inhibition, include metalloprotease inhibitors like 1,10-phenanthroline. Be aware that these zinc chelators may affect other metalloenzymes, so include appropriate controls.
Q7: What are the limitations of current DUB inhibitors? A: Key limitations include:
- Most commercial inhibitors target cysteine proteases, with fewer options for metalloproteases
- Selectivity varies significantly, with many inhibitors affecting multiple DUBs
- Batch-to-batch variability for some inhibitors like PR-619 [14]
- Cellular permeability challenges for in vivo applications
- Potential off-target effects on other cysteine-dependent enzymes
Q8: How can I assess DUB selectivity when using inhibitors? A: Activity-based protein profiling (ABPP) using ubiquitin-based probes provides a powerful method to assess inhibitor selectivity across multiple endogenous DUBs simultaneously in cell lysates [15]. This approach allows screening against 50+ DUBs in a single experiment and can identify off-target effects.
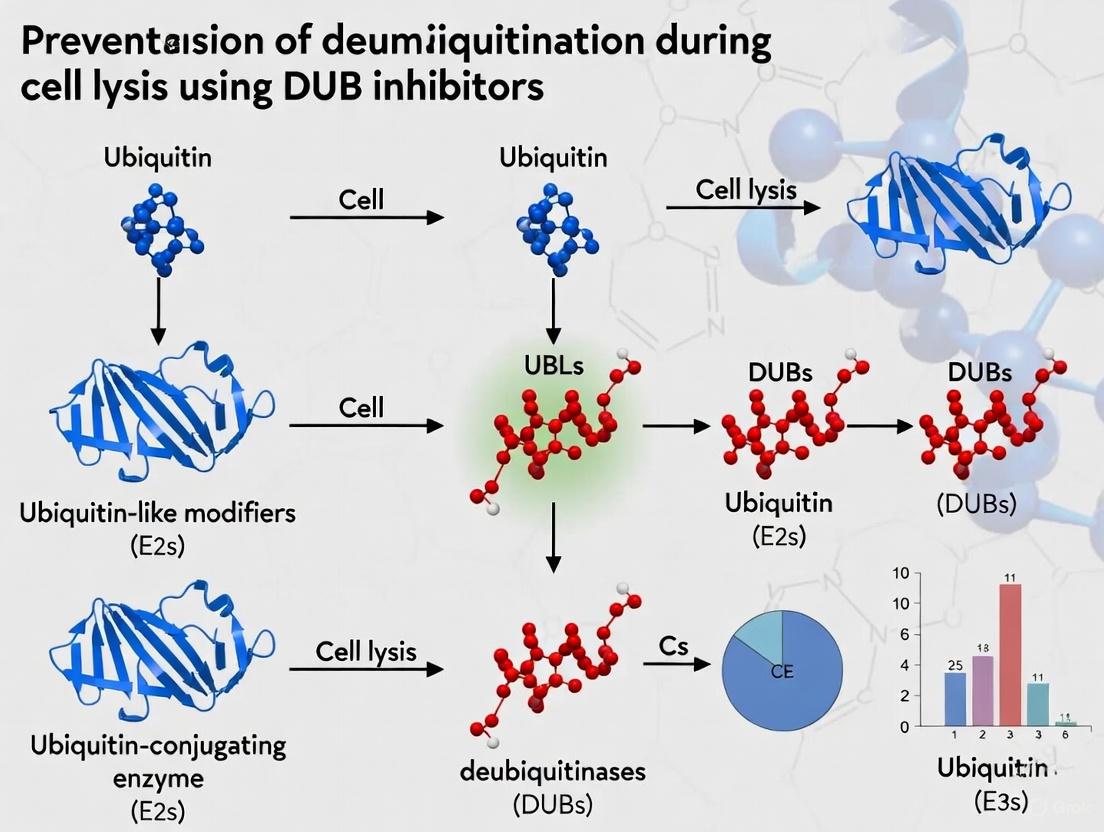
When cells are lysed for experimental analysis, the careful compartmentalization maintained in living cells is abruptly destroyed. This breakdown releases deubiquitinating enzymes (DUBs) from their regulated environments and provides them with artificial access to ubiquitinated substrates from which they would normally be separated. DUBs are cysteine proteases that cleave ubiquitin from protein substrates, thereby reversing ubiquitin signaling and preventing proteasomal degradation [5] [6]. During cell lysis, the sudden mixing of cellular components can trigger artifactual deubiquitination events that compromise experimental results by altering the true ubiquitination status of proteins within living cells. Understanding and controlling this phenomenon is crucial for researchers investigating ubiquitin-dependent processes in signaling, DNA repair, and protein degradation pathways [5].
Key Concepts: DUB Biology and Experimental Artifacts
DUB Families and Mechanisms
Deubiquitinating enzymes comprise approximately 100 proteases in humans, categorized into several subfamilies based on their catalytic domains and mechanisms [16] [6]. The major families include:
- Ubiquitin-Specific Proteases (USPs): The largest subfamily with diverse structural variations
- Ubiquitin C-terminal Hydrolases (UCHs): Involved in ubiquitin precursor processing
- Ovarian Tumor Proteases (OTUs): Known for linkage specificity toward ubiquitin chains
- JAMM/MPN metalloproteases: Zinc-dependent metalloproteases (the only non-cysteine protease DUBs)
- MJD, MINDY, and ZUP1 families
With the exception of JAMM metalloproteases, most DUBs are cysteine proteases that utilize a catalytic triad (Cys, His, Asp/Asn) to cleave isopeptide bonds between ubiquitin and substrate proteins [16]. This catalytic cysteine has a low pKa, making it particularly sensitive to oxidation and other modifications that can affect activity [5].
Compartmentalization Breakdown During Lysis
In intact cells, DUBs and their substrates are strategically localized within different cellular compartments—nucleus, cytoplasm, organelles, and membrane-bound structures. This spatial separation ensures that deubiquitination occurs only at specific times and locations in response to cellular signals. During cell lysis, these physical barriers are disrupted, resulting in:
- Artificial substrate access: DUBs encounter substrates they would not normally access in living cells
- Loss of regulatory complexes: DUBs are separated from binding partners that modulate their activity
- Mixing of cofactors: Cellular reductants and oxidants are redistributed, altering DUB redox states
Vulnerability of the Catalytic Cysteine
The catalytic cysteine residue in DUB active sites is particularly sensitive to oxidative modification. Reactive oxygen species can reversibly inactivate many DUBs by oxidizing this cysteine, abrogating isopeptide-cleaving activity without affecting ubiquitin binding affinity [5]. This redox sensitivity is associated with DUB activation wherein the active site cysteine is converted to a deprotonated state that is prone to oxidation. During cell lysis, changes in the redox environment can significantly impact DUB activity and create experimental artifacts.
Troubleshooting Guide: Common Issues and Solutions
| Problem | Possible Cause | Solution | Verification Method |
|---|---|---|---|
| Unexpected loss of ubiquitin signals | Artifactual DUB activation during lysis | Add DUB inhibitors to lysis buffer; optimize lysis conditions | Compare with/without inhibitors; use multiple ubiquitin antibodies |
| Inconsistent DUB activity measurements | Redox fluctuations affecting catalytic cysteine | Include redox regulators (DTT, GSH) in lysis buffer; work under anaerobic conditions | Measure DUB activity with/without reducing agents |
| Incomplete DUB inhibition | Insufficient inhibitor concentration or specificity | Use combination inhibitors; validate inhibitor efficacy | Test inhibitor concentration series; use activity-based probes |
| Variability between experimental replicates | Inconsistent lysis conditions or timing | Standardize lysis protocol; minimize time between lysis and analysis | Include internal controls; standardize protein quantification |
| Difficulty detecting specific DUB-substrate relationships | Compartmentalization loss allowing non-specific deubiquitination | Use crosslinking before lysis; implement rapid lysis and inhibition | Compare crosslinked vs. non-crosslinked samples |
Frequently Asked Questions (FAQs)
Q1: Why does cell lysis specifically activate DUBs rather than inhibit them? Cell lysis disrupts cellular compartmentalization that normally separates DUBs from their potential substrates. Additionally, lysis alters the redox environment, which can activate certain DUBs by reducing their catalytic cysteine residues. The sudden mixing of cellular contents provides artificial access to substrates that DUBs would not encounter in intact cells [5] [17].
Q2: What are the most critical factors to control during lysis to prevent artifactual deubiquitination? The most critical factors are:
- Temperature: Perform all lysis steps at 4°C to slow enzymatic activity
- Inhibitors: Include DUB inhibitors in your lysis buffer
- Speed: Process samples quickly and consistently
- Redox environment: Control reducing/oxidizing conditions based on your experimental needs
- Standardization: Use consistent lysis conditions across all samples [17] [18]
Q3: How can I verify that my lysis conditions are effectively preserving native ubiquitination states? Several verification approaches include:
- Using activity-based probes to monitor residual DUB activity after lysis
- Comparing time-course experiments to check for progressive loss of ubiquitin signals
- Implementing crosslinking strategies before lysis to capture native interactions
- Testing different inhibitor combinations to identify optimal conditions [18] [15]
Q4: Are certain cell types more susceptible to DUB activation during lysis? Yes, cell types with higher inherent DUB activity or different subcellular organizations may show greater susceptibility. For example, animal cells lyse more readily but may release DUBs more quickly, while plant and bacterial cells with rigid cell walls require more vigorous disruption methods that could potentially activate stress-responsive DUBs [17].
Q5: Can I use the same DUB inhibitors for all DUB families? No, different DUB families have distinct structural features and catalytic mechanisms that require specific inhibitors. Broad-spectrum DUB inhibitors like PR-619 can be useful for initial experiments but may not fully inhibit all DUB classes. For specific research questions, selective inhibitors against particular DUBs (e.g., USP1, UCHL5, or VCPIP1 inhibitors) may be necessary [16] [6] [15].
Experimental Protocols & Methodologies
Standardized Lysis Protocol for Preserving Ubiquitination States
This protocol minimizes artifactual deubiquitination during cell processing:
Reagents Required:
- Lysis buffer (50 mM Tris pH 7.4, 250 mM sucrose, 5 mM MgCl₂, 1 mM ATP)
- DUB inhibitor cocktail (see Section 6 for specific recommendations)
- Protease inhibitors (without DUB inhibitory activity)
- 500 mM DTT stock solution
- N-ethylmaleimide (NEM) for cysteine alkylation
Procedure:
- Pre-chill all equipment and buffers to 4°C
- Prepare fresh lysis buffer supplemented with DUB inhibitors immediately before use
- For adherent cells: Remove media, rinse with cold PBS, and add lysis buffer directly to cells
- For suspension cells: Pellet cells, rinse with cold PBS, and resuspend in lysis buffer
- Incubate on ice for 15-30 minutes with gentle agitation
- For mechanical disruption, use glass beads (1:1 mass:volume ratio) and vortex at maximum agitation for 30 minutes at 4°C [18]
- Clear lysates by centrifugation at 5,030 × g for 5 minutes to remove nuclei and unbroken cells
- Transfer supernatant to fresh tubes and process immediately or flash-freeze in liquid nitrogen
Critical Steps:
- Maintain temperature at 4°C throughout the process
- Include appropriate controls without DUB inhibitors to assess artifact magnitude
- Process all samples consistently with the same lysis duration and conditions
- Use NEM (10-20 mM) after lysis to alkylate free cysteines and "trap" DUBs in their current state [5]
Assessing DUB Activity During Lysis Using Activity-Based Probes
Activity-based probes (ABPs) covalently modify active DUBs and allow direct visualization of their activity status:
Protocol:
- Prepare cell lysates as described above, with and without DUB inhibitors
- Perform bicinchoninic acid (BCA) assay to determine protein concentration
- Incubate 20 μg of total protein with HA-Ub-VS probe (50 nM final concentration) for 1 hour at 37°C [18]
- Stop reaction with Laemmli sample buffer and heat at 95°C for 5 minutes
- Resolve proteins by SDS-PAGE using 4-20% Tris-glycine gels
- Transfer to PVDF membrane and block with 5% milk in PBS
- Incubate with anti-HA primary antibody (1:10,000 dilution) followed by appropriate HRP-conjugated secondary antibody
- Develop blot and compare signal intensity between samples with and without DUB inhibitors
Interpretation: Reduced ABP labeling in inhibitor-treated samples indicates effective DUB inhibition. Persistent labeling suggests incomplete inhibition and potential for artifacts.
Research Reagent Solutions
| Reagent | Function | Application Notes |
|---|---|---|
| PR-619 | Broad-spectrum DUB inhibitor | Useful for initial experiments but lacks specificity; typical working concentration: 10-50 μM |
| N-Ethylmaleimide (NEM) | Cysteine alkylating agent | Irreversibly inactivates cysteine-dependent DUBs; use at 10-20 mM; add after lysis |
| HA-Ub-Vinyl Sulfone (HA-Ub-VS) | Activity-based DUB probe | Covalently labels active DUBs; confirms inhibitor efficacy; use at 50 nM [18] |
| Dithiothreitol (DTT) | Reducing agent | Maintains catalytic cysteine in reduced state; can activate certain DUBs; use at 1-5 mM [5] [18] |
| MG-132 | Proteasome inhibitor | Prevents degradation of deubiquitinated proteins; use at 10-20 μM |
| B-PER Bacterial Protein Extraction Reagent | Specialized lysis reagent | Mild extraction for gram-positive and gram-negative bacteria; includes lysozyme and nuclease [17] |
| Inclusion Body Solubilization Reagent | Denaturing lysis conditions | Useful for studying insoluble ubiquitinated proteins; may require refolding steps [17] |
Signaling Pathways and Workflow Diagrams
Diagram 1: DUB Activation During Cell Lysis: Problem and Prevention Pathways. This workflow illustrates the cascade from cell lysis to experimental artifacts, alongside key prevention strategies.
Diagram 2: Catalytic Cysteine Regulation and Detection. This diagram shows the redox sensitivity of the DUB catalytic cysteine and methods for controlling and detecting its activity state.
| DUB Family | Sensitivity to Redox Changes | Effective Inhibitors | Recommended Lysis Conditions |
|---|---|---|---|
| USP | High (reversible oxidation) [5] | PR-619, specific USP inhibitors [16] | Reducing environment (1-5 mM DTT) + inhibitor cocktail |
| UCH | Moderate to high | Ub-VS derivatives, LDN-57444 [16] | Oxidizing conditions can preserve inactivity; NEM alkylation |
| OTU | Variable | PR-619, specific OTU inhibitors [15] | Test reducing vs. non-reducing conditions empirically |
| JAMM | Low (metalloproteases) | Metal chelators (EDTA, 1,10-phenanthroline) [16] | Metal chelators in lysis buffer |
| MJD | High | Broad-spectrum cysteine inhibitors | Strong reducing agents required |
Advanced Applications: DUB Inhibitors in Research
The development of selective DUB inhibitors has accelerated significantly in recent years, with new compounds emerging against various DUB family members [16] [15]. These inhibitors serve not only as potential therapeutics but also as essential research tools for controlling DUB activity during experimental procedures:
Recent Advances:
- USP1 inhibitors: Target USP1-UAF1 complex for DNA damage response studies
- UCHL5 inhibitors: Investigated for NLRP3 inflammasome regulation and HCV infection [19]
- VCPIP1 inhibitors: New selective probes with nanomolar potency [15]
- Compound screening platforms: Activity-based protein profiling (ABPP) enables high-throughput screening against multiple endogenous DUBs [15]
These advanced chemical tools provide researchers with more specific options for preventing artifactual deubiquitination during cell lysis, moving beyond broad-spectrum approaches to targeted inhibition of specific DUB families implicated in particular experimental systems.
Deubiquitinating enzymes (DUBs) comprise a family of approximately 100 proteases that catalyze the removal of ubiquitin from protein substrates, thereby reversing ubiquitination signals [20] [9]. This dynamic process regulates diverse cellular functions including protein degradation, localization, protein-protein interactions, and signal transduction pathways [21] [9]. In research settings, uncontrolled DUB activity during sample preparation can generate significant experimental artifacts that compromise data interpretation, particularly in protein detection assays and signaling pathway analysis. When cell lysis occurs without adequate DUB inhibition, naturally occurring DUBs remain active and can rapidly deubiquitinate substrates, leading to: (1) loss of biologically relevant ubiquitination signals, (2) misinterpretation of protein regulation mechanisms, and (3) incorrect conclusions about signaling pathway activation states. This technical support article provides comprehensive troubleshooting guidance and validated protocols to prevent these artifacts, ensuring accurate experimental outcomes in ubiquitination-related research.
Troubleshooting Guide: Identifying and Resolving Deubiquitination Artifacts
Weak or Absent Ubiquitin Signal in Western Blotting
| Possible Cause | Recommended Solution | Underlying Principle |
|---|---|---|
| Active DUBs during cell lysis | Add pan-DUB inhibitors (e.g., PR-619) to lysis buffer immediately before use [2] | PR-619 is a broad-spectrum DUB inhibitor that induces ubiquitinated protein accumulation by blocking deubiquitination [2] |
| Insufficient inhibition of DUB activity | Use combination inhibitor approach (e.g., 20µM PR-619 + 10µM PYR-41) [2] | PYR-41 inhibits ubiquitin E1 enzyme, reducing ubiquitin charging and working synergistically with DUB inhibitors [2] |
| Protein degradation during processing | Keep samples on ice, use pre-chilled buffers, and process quickly | DUBs remain active at low temperatures; inhibition is required regardless of temperature control |
| Incompatible lysis buffer composition | Ensure DUB inhibitors are compatible with detergent system; avoid reducing agents that may inhibit certain inhibitors | Some DUB inhibitors rely on cysteine modification and may be compromised by strong reducing agents |
Unusual Banding Patterns in Ubiquitin Detection
| Observed Artifact | Potential Interpretation | Resolution Strategy |
|---|---|---|
| Smearing throughout lanes | Accumulation of heterogeneous ubiquitinated species | Optimize inhibitor concentration; confirm efficacy using positive controls |
| Loss of high-molecular-weight ubiquitin conjugates | Excessive DUB activity preferentially removing polyubiquitin chains | Use fresh DUB inhibitors; avoid freeze-thaw cycles of inhibitor stocks |
| Extra bands at unexpected molecular weights | Non-specific antibody binding or protein degradation | Include control without DUB inhibitor to distinguish specific ubiquitination patterns |
| Complete absence of signal | Over-blocking or antigen masking | Compare different blocking agents (BSA vs. non-fat milk); optimize antibody concentrations [22] |
Experimental Protocols: Preserving Ubiquitination States During Sample Preparation
Optimized Cell Lysis Protocol with DUB Inhibition
Purpose: To effectively extract proteins while preserving ubiquitination states by inhibiting endogenous DUB activity.
Materials Needed:
- Pan-DUB inhibitor (e.g., PR-619 at 50-100mM stock in DMSO)
- Ubiquitin E1 inhibitor (e.g., PYR-41 at 10mM stock in DMSO) [2]
- Complete protease inhibitor cocktail (without DUB inhibitors)
- Lysis buffer (e.g., RIPA or NP-40 based)
- Pre-chilled PBS
- Cell scrapers (for adherent cells)
Procedure:
- Prepare fresh working inhibitor solution by diluting PR-619 to 20µM and PYR-41 to 10µM in lysis buffer.
- Aspirate culture media from cells and wash once with ice-cold PBS.
- Add appropriate volume of inhibitor-containing lysis buffer to cells (e.g., 100-200µL for a 6-well plate).
- Incubate on ice for 5-10 minutes with occasional rocking.
- Scrape adherent cells thoroughly and transfer lysate to pre-chilled microcentrifuge tube.
- Clarify by centrifugation at 12,000 × g for 10 minutes at 4°C.
- Transfer supernatant to new tube and proceed immediately to protein quantification or store at -80°C.
Validation: Confirm efficacy of DUB inhibition by comparing with samples lysed without DUB inhibitors, monitoring accumulation of high-molecular-weight ubiquitinated proteins [2].
Activity-Based Profiling of DUB Inhibition Efficacy
Purpose: To validate the effectiveness of DUB inhibition protocols using activity-based probes.
Materials Needed:
- Ubiquitin-based active site-directed probes (Ub-VME or Ub-PA) [23]
- Cell lysates prepared with and without DUB inhibitors
- Streptavidin beads (for pull-down)
- SDS-PAGE equipment
- Anti-ubiquitin antibodies
Procedure:
- Incubate cell lysates (10-20µg) with Ub-VME or Ub-PA probes (1µM) for 30 minutes at 37°C [23].
- Stop reaction with SDS sample buffer.
- Separate proteins by SDS-PAGE and transfer to membrane.
- Detect labeled DUBs using streptavidin-HRP or anti-ubiquitin antibodies.
- Compare labeling intensity between samples prepared with and without DUB inhibitors.
Interpretation: Effective DUB inhibition should show reduced probe labeling in inhibitor-treated samples, confirming DUB inactivation during lysis.
The Scientist's Toolkit: Essential Reagents for Controlling Deubiquitination
Research Reagent Solutions
| Reagent/Category | Specific Examples | Mechanism of Action | Application Notes |
|---|---|---|---|
| Broad-Spectrum DUB Inhibitors | PR-619 | Pan-DUB inhibitor inducing ubiquitin-protein aggregation [2] | Use at 20-50µM in lysis buffer; compatible with various detection methods |
| E1 Ubiquitin Activating Enzyme Inhibitors | PYR-41 | Inhibits ubiquitin activation, reducing substrate ubiquitination [2] | Use at 10µM in combination with DUB inhibitors for enhanced effect |
| Activity-Based Probes | Ub-VME, Ub-PA | Covalently label active site cysteine of DUBs [23] | Essential for validating DUB inhibition efficacy; use at 1µM concentration |
| Selective DUB Inhibitors | VLX1570 (targets USP14/UCHL5) [24] | Specific inhibition of proteasome-associated DUBs | Useful for studying specific DUB functions; limited for general lysis protection |
| Ubiquitin Chain Reference Standards | K48-linked, K63-linked di-Ub/tetra-Ub chains [23] | Linkage-specific ubiquitin standards | Critical controls for assessing DUB activity and linkage specificity |
FAQs: Addressing Common Technical Challenges
Q1: Why do I still detect background DUB activity even when using recommended DUB inhibitors?
A: Persistent DUB activity typically results from: (1) insufficient inhibitor concentration - perform dose optimization for your specific system; (2) incomplete inhibition of all DUB classes - consider combining inhibitors with different specificities; (3) inhibitor degradation - prepare fresh stocks and avoid multiple freeze-thaw cycles; or (4) lysis buffer incompatibility - ensure detergent system doesn't interfere with inhibitor function. Validation with activity-based probes is recommended [23].
Q2: How does uncontrolled deubiquitination specifically affect interpretation of Wnt signaling pathways?
A: In Wnt signaling, ubiquitination directly regulates key components including β-catenin, Axin, GSK3, and Dvl [21]. Unchecked deubiquitination during sample preparation can: (1) artificially stabilize β-catenin, leading to false conclusions about pathway activation; (2) alter the degradation kinetics of pathway regulators; and (3) obscure phosphorylation-dependent ubiquitination events that are crucial for pathway regulation. These artifacts fundamentally compromise mechanistic studies of Wnt signaling modulation.
Q3: What are the best practices for storing and handling DUB inhibitors to maintain efficacy?
A: Follow these guidelines: (1) aliquot inhibitors in single-use volumes to avoid freeze-thaw cycles; (2) store at -80°C in anhydrous DMSO; (3) protect from light and moisture; (4) add inhibitors to lysis buffer immediately before use; and (5) avoid extended storage of inhibitor-containing buffers even at -20°C. Periodically validate inhibitor efficacy using activity-based probes [23].
Q4: Can I use the same DUB inhibition strategy for all cell types and tissues?
A: While the fundamental principles apply universally, optimization may be required for: (1) tissues with high intrinsic DUB activity (e.g., brain tissue); (2) cells expressing unusual DUB profiles (e.g., cancer cells with DUB amplification); and (3) subcellular fractionation studies where compartment-specific DUBs may be enriched. Always validate your inhibition strategy for each novel experimental system.
Advanced Applications: DUB Inhibitors in Cancer Research and Therapeutic Development
The controlled inhibition of DUBs has significant implications beyond preventing experimental artifacts, particularly in cancer research and drug development. Many DUBs are genetically altered or dysregulated in various cancers, functioning as either oncogenes or tumor suppressors [20] [24]. For instance, USP6 overexpression due to chromosomal rearrangements drives aneurysmal bone cysts, while CYLD mutations are associated with familial cylindromatosis [20]. The pan-DUB inhibitor PR-619 has demonstrated profound anti-cancer effects in oesophageal squamous cell carcinoma, inducing G2/M cell cycle arrest, apoptosis, and autophagy through ubiquitin-protein aggregation-activated ER stress [2]. Several DUB-targeting therapeutics have entered clinical development, including VLX1570 (targeting USP14/UCHL5) for multiple myeloma and KSQ-4279 for solid tumors [24]. These developments highlight the dual importance of DUB inhibition: as a crucial methodological approach for accurate research and as a promising therapeutic strategy.
The Scientist's Toolkit: Research Reagent Solutions
Table 1: Essential Reagents for DUB Research
| Reagent Name | Function/Application | Key Features |
|---|---|---|
| Biotin-UbVMe [12] | Activity-based DUB probe; covalently binds active site cysteine of DUBs. | Contains N-terminal Avi-Tag for biotinylation, C-terminal vinyl methyl ester (VME) electrophile; used for enrichment and detection. |
| Ub-AMC / Ub-Rho110 [12] | Fluorogenic DUB substrates for enzymatic activity assays. | DUB cleavage releases fluorescent AMC or Rhodamine 110; Ub-Rho110 offers red-shifted spectra, reducing compound interference. |
| DUB-Glo Assay [12] | Bioluminescent assay for DUB activity. | Offers low background signal, suitable for high-throughput screening (HTS) campaigns. |
| MLN4924 [25] | Inhibitor of NEDD8-activating E1 enzyme. | Indirectly affects a subset of Cullin-RING E3 ubiquitin ligases; used as a control or tool compound. |
| Auranofin [25] | Inhibitor of proteasome-associated DUBs UCHL5 and USP14. | Used to study the role of 19S proteasome-associated DUBs in cancer cell survival. |
| PR-619 / HBX41108 [26] | Broad-spectrum, covalent DUB inhibitors. | Useful as positive controls in activity assays and for validating DUB-dependent cellular phenomena. |
| XL177A [26] | Selective USP7 inhibitor. | Example of a selective chemical probe; used to interrogate specific DUB biology. |
| SB1-F-22 (N-cyanopyrrolidine) [26] | Covalent inhibitor targeting UCHL1 active site cysteine. | Represents a chemotype inspired by patent literature; used for UCH-family DUB targeting. |
Troubleshooting Guides & FAQs
FAQ 1: Why is preventing deubiquitination during cell lysis critical for my experiments, and how can I achieve it?
Answer: Preventing deubiquitination during cell lysis is essential to preserve the in vivo ubiquitination status of your proteins of interest. DUBs remain active in cell lysates and can rapidly remove ubiquitin chains from substrates after lysis, leading to inaccurate representation of protein stability, degradation, and signaling events [12]. This is a fundamental consideration for any research framed within the context of investigating DUB functions.
Solution:
- Use Comprehensive DUB Inhibitors: Add a cocktail of broad-spectrum DUB inhibitors to your lysis buffer immediately upon cell disruption. Common choices include:
- PR-619: A cell-permeable, pan-DUB inhibitor effective in the low micromolar range.
- N-Ethylmaleimide (NEM): An alkylating agent that inhibits cysteine proteases, including most DUB families.
- Optimize Buffer Conditions: Keep lysates on ice and perform procedures rapidly to minimize enzymatic activity. The use of these inhibitors in lysis buffers is a standard practice for assays like the cell lysate-based AlphaLISA, which relies on capturing the true state of DUB activity and ubiquitination [12].
FAQ 2: My high-throughput screen against a DUB yielded numerous hits, but I suspect many are non-selective or false positives. How can I triage them effectively?
Answer: This is a common challenge in DUB drug discovery. Early-generation DUB inhibitors are often multitargeted, and false positives can arise from compound interference (e.g., fluorescence, aggregation) [26].
Solution: Implement an Orthogonal Assay Cascade:
- Primary Screening: Use a robust, physiologically relevant primary screen. The cell lysate-based ABPP (Activity-Based Protein Profiling) platform is highly recommended. It screens compounds against endogenous, full-length DUBs in their native cellular environment, providing selectivity data across many DUBs simultaneously [26].
- Orthogonal Validation: Confirm hits from your primary screen using a different assay technology.
- Fluorometric Assays: Use Ub-AMC or Ub-Rho110 cleavage assays with the purified recombinant DUB to confirm direct enzymatic inhibition [12].
- Gel-Based Assays: Employ western blotting to monitor stabilization of known ubiquitinated substrates of the DUB in cells treated with your hit compounds.
- Counter-Screening: Test your hits against a panel of other DUBs and related enzymes (e.g., other cysteine proteases) to define selectivity. The ABPP platform is ideal for this, as it can profile compound selectivity against dozens of endogenous DUBs in a single experiment [26].
FAQ 3: I am struggling to obtain sufficient quantities of active, recombinant DUB protein for biochemical assays. What are my options?
Answer: Many human DUBs are large, multi-domain proteins that are challenging to express and purify in active form in sufficient quantities for high-throughput screening (HTS) [12].
Solution:
- Consider Alternative Expression Systems: If E. coli expression fails, try baculovirus-mediated expression in insect cells, which often better handles the folding and post-translational modifications of complex human proteins.
- Utilize Cell Lysate-Based Assays: Shift your strategy to a platform that does not require purified protein. The AlphaLISA DUB assay and the ABPP platform are designed to work with DUBs expressed in and extracted from human cells. This not only circumvents the purification problem but also recapitulates a more physiologically relevant environment, as many DUBs require binding partners for full activity and specificity [12] [26].
- Focus on Catalytic Domains: For initial mechanistic or structural studies, consider expressing and purifying only the well-characterized catalytic domain of the DUB, which is often more tractable.
FAQ 4: How can I determine if a DUB is a realistic therapeutic target for a specific cancer or neurodegenerative disease?
Answer: Target validation requires demonstrating that the DUB is functionally involved in a disease-relevant pathway and that its inhibition has a therapeutic effect.
Solution: A Multi-Faceted Validation Approach:
- Genetic Evidence: Use siRNA, shRNA, or CRISPR-Cas9 to knock down or knock out the DUB in disease-relevant cell models (e.g., cancer cell lines, neuronal models). Look for phenotypic changes such as reduced cell proliferation, induction of apoptosis, or decreased levels of pathogenic proteins (e.g., α-synuclein, Tau) [27] [28].
- Clinical Correlation: Analyze human tissue databases (e.g., The Cancer Genome Atlas) for evidence of DUB dysregulation (overexpression, amplification, mutation) in patient tumors and correlate this with clinical outcomes [27] [25].
- Chemical Probe Validation: Use a selective, well-characterized small-molecule inhibitor (chemical probe) of the DUB. The phenotypic effects of genetic knockdown should be recapitulated by pharmacological inhibition. The development of probes for DUBs like USP7, USP28, and VCPIP1 provides a blueprint for this approach [26].
- Mechanistic Insight: Identify the key substrates stabilized by the DUB (e.g., MYC, HIF1α stabilized by USP29 [27] or pathogenic proteins in NDs [28]). Rescuing the phenotype by reconstituting the substrate can confirm the mechanism.
Experimental Protocol: Cell Lysate-Based DUB Inhibitor Screening via ABPP
This protocol outlines a method for screening compounds for DUB inhibition using endogenous DUBs in cell lysates, leveraging the power of Activity-Based Protein Profiling (ABPP) and quantitative mass spectrometry [26].
1. Reagent Preparation:
- Biotinylated Ubiquitin Probes: Prepare a 1:1 mixture of biotin-Ub-VME (vinyl methyl ester) and biotin-Ub-PA (propargylamide). These probes covalently label the active site cysteine of most DUBs [12] [26].
- Compound Library: Dissolve compounds in DMSO. A final concentration of 50 µM is typical for a primary screen.
- Cell Lysate: Harvest HEK293 or other relevant cells. Lyse cells in a appropriate buffer (e.g., 50 mM Tris pH 8.0, 150 mM NaCl, 0.5% NP-40) supplemented with broad-spectrum DUB inhibitors. Clear the lysate by centrifugation. Determine protein concentration.
2. Primary Screening Incubation:
- In a 96-well plate, mix 50 µg of cell lysate per well with 1 µL of compound (or DMSO for controls).
- Incubate for 30 minutes at room temperature to allow compound binding.
- Add the biotin-Ub probe mixture (final concentration ~100 nM) and incubate for an additional 60 minutes. The probe will label all DUBs not blocked by a bound inhibitor.
3. Sample Processing for Mass Spectrometry:
- Denature the samples and digest with trypsin.
- Label the peptides from different samples with isobaric TMT (Tandem Mass Tag) reagents. This allows for multiplexing (e.g., 16-plex) and relative quantification across samples.
- Enrich biotinylated peptides (which contain the DUB-derived peptides covalently linked to the probe) using streptavidin beads.
4. Data Acquisition and Analysis:
- Analyze the enriched peptides by nanoflow liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS).
- Use database searching to identify the DUB peptides. The TMT reporter ion intensities for each DUB peptide are proportional to the amount of probe labeling, and thus inversely proportional to compound inhibition.
- A "hit compound" is typically defined as one that reduces ABP labeling of a specific DUB by ≥50% compared to the DMSO control [26].
DUB Signaling Pathways and Experimental Workflows
Diagram: DUB Regulation of Key Oncogenic Pathways
Diagram: Workflow for DUB Inhibitor Discovery & Validation
Practical Implementation of DUB Inhibitors in Cell Lysis Protocols
Deubiquitinating enzymes (DUBs) represent a family of approximately 100 proteases that catalyze the removal of ubiquitin from protein substrates, thereby reversing the activity of E3 ubiquitin ligases and playing central roles in regulating cellular processes such as protein degradation, DNA repair, and cell signaling [29] [20]. The pharmacologic interrogation of this important protein family has been hampered by a historical lack of selective chemical probes, impeding both basic research and therapeutic development [29]. DUB inhibitors have emerged as critical tools for disrupting deubiquitination processes, with applications ranging from fundamental mechanism discovery to potential cancer therapeutics [20].
Two primary classes of DUB inhibitors have been developed: broad-spectrum inhibitors that simultaneously target multiple DUBs, and selective inhibitors that specifically target individual DUB family members. Each class possesses distinct characteristics, applications, and limitations that researchers must consider when designing experiments. Broad-spectrum inhibitors like PR-619 provide valuable tools for initial discovery research and for preserving ubiquitinated proteins during cell lysis, while selective inhibitors such as ML323 (targeting USP1-UAF1) and XL177A (targeting USP7) enable precise pharmacological interrogation of specific DUB functions [30] [31] [32].
The following sections provide a comprehensive technical resource for researchers working with DUB inhibitors, including comparative characterization data, experimental protocols, troubleshooting guidance, and reagent information specifically framed within the context of preventing deubiquitination during cell lysis and advancing DUB-targeted therapeutic discovery.
DUB Inhibitor Classes: Characteristics and Applications
Comparative Analysis of Inhibitor Classes
Table 1: Characteristics of Broad-Spectrum vs. Selective DUB Inhibitors
| Characteristic | Broad-Spectrum Inhibitors | Selective Inhibitors |
|---|---|---|
| Target Range | Multiple DUBs across subfamilies (e.g., PR-619 inhibits many cysteine-reactive DUBs) [30] | Individual DUBs or specific complexes (e.g., ML323 specifically inhibits USP1-UAF1; XL177A targets USP7) [31] [32] |
| Primary Applications | - Preservation of ubiquitinated proteins during cell lysis [30]- Initial screening and phenotyping- Studying global ubiquitination dynamics | - Precise target validation- Therapeutic development- Mapping specific DUB-substrate relationships [32] |
| Common Examples | PR-619 (reversible, 5-20 μM IC₅₀ range) [30] | ML323 (USP1-UAF1, nM potency), XL177A (USP7, 0.34 nM IC₅₀) [31] [32] |
| Key Advantages | - Immediate effects on global ubiquitination- Useful for pathway discovery- Cost-effective for initial studies | - Minimal off-target effects- Clear interpretation of results- Better therapeutic potential |
| Major Limitations | - Difficult to attribute effects to specific DUBs- Potential compensatory mechanisms- Higher risk of cellular toxicity | - Require prior knowledge of target- More resource-intensive development- Limited for complex polygenic diseases |
| Recommended Use Cases | - Lysis buffer additive (50-100 μM) to preserve ubiquitination [30]- Initial studies of DUB involvement in processes- Identifying DUB-sensitive cellular processes | - Validating individual DUB functions- Probe development for specific DUBs- Targeted therapeutic applications |
Molecular Mechanisms and Structural Basis
The structural basis for DUB inhibitor selectivity stems from the diverse active site architectures across DUB subfamilies. Broad-spectrum inhibitors typically target conserved catalytic cysteine residues found in multiple DUB families, while selective inhibitors exploit unique structural features surrounding individual DUB active sites. For instance, the broad-spectrum inhibitor PR-619 contains thiocyanate groups that react with catalytic cysteine residues across numerous cysteine protease DUB families [30].
In contrast, selective inhibitors achieve their specificity through optimized interactions with unique structural elements. The development of selective inhibitors has been accelerated by structure-guided approaches that analyze DUB-ligand and DUB-ubiquitin co-structures to identify regions around the catalytic site that favor compound interaction and potential selectivity determinants [29]. For example, the selective USP1-UAF1 inhibitor ML323 achieves its exceptional selectivity profile by specifically engaging with unique structural features of the USP1-UAF1 complex rather than simply targeting the conserved catalytic domain [31].
Recent advances in rational library design have embraced this structural complexity through chemical diversification strategies that incorporate noncovalent building blocks, linkers, and electrophilic warheads designed to interact with both conserved and unique regions around DUB catalytic sites [29]. This approach has successfully yielded selective inhibitors for previously untargeted DUBs, demonstrating the feasibility of developing selective compounds across this important gene family.
Experimental Protocols & Methodologies
Protocol for Cell Lysis with DUB Inhibition
Purpose: To effectively preserve ubiquitin conjugates during cell extraction by inhibiting endogenous deubiquitinating enzymes.
Equipment:
- Sonicator or vortexer
- Micropipette
- Conical tube (sized accordingly for cell sample)
- Microcentrifuge tube (sized accordingly for cell sample)
- Microcentrifuge
- Cell scraper or spatula [33]
Reagents:
- Mammalian cells grown in adherent culture or suspension
- Ice-cold RIPA Lysis Buffer or other appropriate lysis buffer
- Protease and Phosphatase Inhibitor Cocktail (100X)
- DUB inhibitor (e.g., PR-619 or selective inhibitors)
- Ice-cold PBS [33]
Procedure:
Preparation of Lysis Buffer: Add 0.1 mL of protease and phosphatase inhibitors to 10 mL RIPA buffer. Add DUB inhibitor to appropriate concentration (e.g., 50-100 μM for PR-619) [30] [33].
Cell Preparation:
- For adherent cells: Culture to ~80% confluence. Aspirate media and keep plate on ice. Wash cell monolayer gently with 10 ml ice-cold PBS. Aspirate excess PBS [33].
- For suspension cells: Culture to 1-2 x 10⁶ cells/ml. Pellet cells at 300 × g for 5 minutes at room temperature. Aspirate media and keep cells on ice. Wash pellet with 5-10 ml ice-cold PBS. Centrifuge again at 300 × g for 5 minutes and aspirate supernatant [33].
Lysis:
Incubation: Incubate lysate on ice for 15 minutes [33].
Homogenization: Sonicate or vortex the lysate three times for 2 seconds each. Rest the lysate at least one minute between pulses. Repeat if lysate remains viscous [33].
Secondary Incubation: Incubate lysate on ice for additional 15 minutes [33].
Clarification: Centrifuge lysate at 13,000 × g for 5 minutes at 4°C [33].
Collection: Transfer supernatant to new microcentrifuge tubes, avoiding disruption of pellet [33].
Storage: Aliquot and store lysate at -20°C for short-term use or -80°C for long-term storage [33].
Validation: Determine protein concentration using Bradford, BCA, or other appropriate protein assay before proceeding to downstream applications [33].
Workflow for DUB Substrate Identification Using Selective Inhibitors
Diagram 1: Proteomics workflow for DUB substrate identification using selective inhibitors. This approach enables comprehensive mapping of DUB substrates by monitoring protein stabilization/destabilization following targeted DUB inhibition [32].
High-Throughput Screening Protocol for DUB Inhibitor Discovery
Purpose: To identify selective DUB inhibitors through high-throughput screening of compound libraries against recombinant DUB enzymes.
Equipment:
- FPLC system with size exclusion chromatography
- Sonicator
- Centrifuge
- Microplate reader for fluorescence-based assays
- Liquid handling robotics for HTS (optional)
Reagents:
- DUB expression vectors (pET28 with 6xHis tag or pGEX6P1 with GST tag)
- BL21(DE3) Competent E. coli
- LB medium with appropriate antibiotics
- IPTG for protein induction
- Lysis buffer (25 mM Tris, pH 8, 10 mM β-mercaptoethanol, 1 M NaCl)
- Ni-NTA Agarose (for 6xHis-tagged proteins) or Glutathione Agarose (for GST-tagged proteins)
- Elution buffers with imidazole (6xHis-tagged) or GST 3C protease (GST-tagged)
- Size exclusion column buffer (25 mM HEPES, pH 7.5, 200 mM NaCl, 1 mM DTT)
- Ubiquitin-Rhodamine110 (Ub-Rho) substrate
- Test compounds in DMSO [34]
Procedure:
Protein Expression:
- Transform BL21(DE3) E. coli with DUB plasmid
- Inoculate single colony into 5 mL LB medium with antibiotic, grow at 37°C for 16-18h
- Dilute culture into 1L LB medium, grow until OD₆₀₀ reaches 0.8-1.0
- Induce protein expression with 100 mg/L IPTG, incubate at 16°C for 18-24h
- Pellet cells by centrifugation at 4,540 × g for 20min at 4°C [34]
Protein Purification:
- Resuspend cell pellet in 50-100mL lysis buffer, stir at 4°C for 30min
- Add PMSF to 10 μg/mL, lyse by sonication on ice
- Centrifuge lysate at 30,000 × g for 40min at 4°C
- Equilibrate appropriate affinity resin in lysis buffer
- Incubate cell lysate with resin at 4°C for 2-4h with gentle agitation
- Wash resin with wash buffer (25 mM Tris, pH 8, 10 mM β-mercaptoethanol, 1 M NaCl, 25 mM imidazole for 6xHis-tagged proteins)
- Elute protein with elution buffer (containing 300 mM imidazole for 6xHis-tagged proteins or GST 3C protease for GST-tagged proteins)
- Concentrate protein using centrifugal filter units
- Further purify via FPLC size exclusion chromatography [34]
Enzyme Activity Assay:
- Prepare reaction buffer optimized for each DUB
- Incubate purified DUB with Ub-Rho substrate in presence of test compounds
- Monitor fluorescence increase (excitation 485 nm, emission 535 nm) over time
- Calculate inhibition relative to DMSO controls [34]
Hit Validation:
Troubleshooting Guides & FAQs
Frequently Asked Questions
Q1: Why should I include DUB inhibitors in my cell lysis buffer, and which type should I choose?
A: Deubiquitinating enzymes remain active during cell lysis and can rapidly remove ubiquitin from proteins of interest, potentially obscuring detection of physiologically relevant ubiquitination events. Including DUB inhibitors in lysis buffers preserves ubiquitin conjugates that would otherwise be lost. For general ubiquitin preservation, broad-spectrum inhibitors like PR-619 (50-100 μM) are recommended. For studies focused on specific DUB substrates, consider including selective inhibitors targeting highly active DUBs in your system, if available [30].
Q2: What is the difference between reversible and irreversible DUB inhibitors?
A: Reversible inhibitors (e.g., PR-619) bind non-covalently to DUBs and their effects can be diluted or reversed. Irreversible inhibitors (e.g., XL177A) typically form covalent bonds with catalytic cysteine residues, resulting in permanent enzyme inhibition. Reversible inhibitors are often preferred for acute treatments, while irreversible inhibitors can provide prolonged inhibition but may require careful concentration optimization to minimize off-target effects [30] [32].
Q3: How do I validate the selectivity of a DUB inhibitor for my study?
A: Several approaches can assess inhibitor selectivity: (1) In vitro profiling against panels of recombinant DUBs; (2) Cellular chemoproteomic methods like activity-based protein profiling (ABPP) that assess engagement of endogenous DUBs; (3) Monitoring changes in known substrate ubiquitination status; (4) Genetic validation using DUB knockout or knockdown cells. For comprehensive selectivity assessment, ABPP platforms can evaluate compound activity against 65+ endogenous DUBs simultaneously [29] [32].
Q4: Can DUB inhibitor treatment itself affect cellular ubiquitin levels?
A: Yes, particularly with broad-spectrum inhibitors. Treatment with non-selective DUB inhibitors like PR-619 causes accumulation of ubiquitinated proteins by blocking the recycling of ubiquitin. This can potentially deplete free ubiquitin pools over extended treatments and cause secondary effects. Selective inhibitors typically have minimal impact on global ubiquitination, making them preferable for prolonged treatments [30].
Q5: What cellular phenotypes should I expect after DUB inhibition?
A: This depends on the specific DUB targeted. Inhibition of DUBs regulating stability of oncoproteins or tumor suppressors may affect cell proliferation and survival. Inhibition of DUBs involved in DNA damage response (e.g., USP1) can sensitize cells to genotoxic agents. Broad-spectrum inhibition typically causes accumulation of polyubiquitinated proteins and may activate stress pathways. Phenotypes should be interpreted cautiously with non-selective inhibitors, as effects may result from combined inhibition of multiple DUBs [31] [20].
Troubleshooting Common Experimental Issues
Table 2: Troubleshooting DUB Inhibitor Experiments
| Problem | Potential Causes | Solutions |
|---|---|---|
| Poor preservation of ubiquitinated proteins during lysis | - Inadequate DUB inhibitor concentration- Insufficient inhibition speed- Improper lysis buffer composition | - Increase DUB inhibitor concentration (e.g., 100 μM PR-619) [30]- Ensure rapid inhibition by adding inhibitors directly to lysis buffer- Include complementary protease inhibitors |
| High background in ubiquitin detection | - Non-specific antibody binding- Incomplete lysate clarification- Protein overloading | - Optimize antibody concentrations- Increase centrifugation speed/time for better clarification- Reduce protein loading; validate with positive/negative controls |
| Cellular toxicity with inhibitor treatment | - Off-target effects- Excessive inhibitor concentration- Prolonged exposure | - Titrate inhibitor to find minimum effective concentration- Use selective inhibitors instead of broad-spectrum- Shorten treatment duration |
| Lack of expected phenotype with selective inhibitor | - Inadequate target engagement- Compensation by related DUBs- Incorrect biological hypothesis | - Verify target engagement using cellular assays- Test combination of inhibitors targeting related DUBs- Validate with genetic approaches (e.g., CRISPR) |
| Inconsistent results between experiments | - Inhibitor stability issues- Variable cell culture conditions- Differences in lysis efficiency | - Prepare fresh inhibitor stocks; avoid freeze-thaw cycles- Standardize cell culture and treatment conditions- Monitor lysis efficiency visually and by protein quantification |
Research Reagent Solutions
Table 3: Essential Reagents for DUB Inhibition Studies
| Reagent/Category | Specific Examples | Key Applications & Functions | Considerations |
|---|---|---|---|
| Broad-Spectrum DUB Inhibitors | PR-619 [30] | - Preserve ubiquitinated proteins during cell lysis (50-100 μM) [30]- Initial studies of DUB involvement in cellular processes | - Reversible inhibitor- Use for acute treatments- Can affect global ubiquitination |
| Selective DUB Inhibitors | ML323 (USP1-UAF1) [31]XL177A (USP7) [32] | - Precise target validation- Therapeutic mechanism studies- Mapping specific DUB substrates [31] [32] | - Validate selectivity for your system- Titrate for optimal concentration- Consider covalent vs. reversible mechanism |
| Activity-Based Probes | Biotin-Ub-VME/Biotin-Ub-PA [29] | - Assess endogenous DUB activity- Profiling inhibitor selectivity- DUB discovery [29] | - Can be combined for broader DUB coverage- Use with quantitative mass spectrometry |
| Lysis & Stabilization Reagents | RIPA Buffer [33]Protease Inhibitor Cocktails [33] | - Maintain protein integrity during extraction- Prevent non-specific proteolysis- Preserve post-translational modifications | - Include DUB inhibitors specifically for ubiquitin preservation- Keep samples cold throughout processing |
| Detection Reagents | Anti-ubiquitin antibodiesTMT multiplexed reagents [32] | - Detect ubiquitinated proteins- Quantitative proteomics for substrate identification [32] | - Validate antibody specificity- Use appropriate multiplexing design for statistical power |
DUB Signaling Pathways in Cancer
Diagram 2: USP1-UAF1 signaling in DNA damage response and chemosensitization. ML323 inhibition of USP1-UAF1 prevents deubiquitination of key DNA repair proteins, enhancing cisplatin sensitivity in cancer cells [31].
The strategic application of both broad-spectrum and selective DUB inhibitors provides powerful approaches for advancing our understanding of deubiquitination biology and developing novel therapeutic strategies. Broad-spectrum inhibitors remain invaluable tools for initial discovery research and for preserving ubiquitination signatures during cell lysis, while selective inhibitors enable precise dissection of individual DUB functions and target validation. The continuing development of increasingly selective chemical probes, coupled with advanced screening platforms such as activity-based protein profiling, is rapidly accelerating pharmacological interrogation of this important gene family. As the field progresses, the appropriate selection and application of these inhibitor classes—with careful consideration of their distinct characteristics and limitations—will be essential for designing robust experiments and generating meaningful biological insights with potential therapeutic applications in cancer and other diseases.
Frequently Asked Questions (FAQs)
FAQ 1: What is the core mechanism of action of the pan-DUB inhibitor PR-619? PR-619 is a broad-spectrum, cell-permeable inhibitor that targets the active site cysteine of cysteine protease DUBs, including USP, UCH, MJD, OTU, and MINDY families [16] [35]. Its primary action is to induce the accumulation of ubiquitinated proteins by blocking deubiquitinating activity. This accumulation can trigger downstream cellular events, most notably Endoplasmic Reticulum (ER) stress, which then activates apoptosis and autophagy pathways [2].
FAQ 2: How can I confirm that PR-619 is working in my cell lysate experiment? The most direct method is to detect the increase in global ubiquitin conjugates via western blotting. Use an anti-ubiquitin antibody to compare lysates treated with PR-619 against a DMSO vehicle control. You should observe a characteristic smear of high-molecular-weight proteins in the treated sample, indicating successful inhibition of DUBs and the accumulation of poly-ubiquitinated substrates [14] [36].
FAQ 3: What is a typical working concentration for PR-619 in cell-based assays? Effective concentrations can vary based on cell type and treatment duration. The table below summarizes concentrations used in published studies.
| Cell Type/Model | Typical Concentration Range | Key Observed Effect | Source |
|---|---|---|---|
| Oesophageal squamous cell carcinoma | 20 - 40 µM | Induced apoptosis & autophagy [2] | |
| Renal fibrosis (mouse model, in vivo) | 100 µg per dose (daily IP injection) | Suppressed renal fibrosis [37] | |
| Retinal ganglion cells (in vivo) | 8.23 µM (intravitreal injection) | Enhanced mitophagy, neuroprotection [38] | |
| U2OS cells (ubiquitinome study) | 10 - 50 µM | Global accumulation of ubiquitin substrates [14] |
FAQ 4: Why does PR-619 treatment lead to autophagy in some cellular models? The induction of autophagy is often a secondary consequence of cellular stress. PR-619-induced ubiquitinated protein aggregates activate ER stress, leading to an increase in cytosolic Ca²⁺ levels. This calcium release activates the CaMKKβ-AMPK signaling pathway, which is a key positive regulator of autophagy [2].
FAQ 5: Are the effects of PR-619 reversible? The inhibition is not considered easily reversible because PR-619 acts as a covalent modifier of the active site cysteine in target DUBs [35]. However, the cellular ubiquitin landscape can recover over time as the inhibitor is cleared and new DUB proteins are synthesized. One study showed that the bulk of ubiquitin conjugates accumulated by DUB inhibition were turned over within 3 hours after co-treatment with a ubiquitin E1 inhibitor (TAK243), which blocks new ubiquitination [14].
Troubleshooting Guide
Problem 1: Excessive Cell Death or Unintended Apoptosis
- Potential Cause: The concentration of PR-619 is too high for your specific cell type, leading to an overwhelming ER stress response and rapid induction of apoptosis [2].
- Solutions:
- Perform a dose-response curve to determine the optimal concentration that achieves DUB inhibition without excessive toxicity. Start testing in the range of 10-50 µM.
- Shorten the treatment duration. Instead of continuous exposure, consider a pulse-treatment followed by a recovery period in fresh medium.
- Use an orthogonal assay (e.g., caspase activity assay) to quantify apoptosis and better define the therapeutic window for your experiment.
Problem 2: No Apparent Increase in Ubiquitin Smear on Western Blot
- Potential Cause 1: Inefficient cell lysis or deubiquitination occurring during lysate preparation.
- Solution: Incorporate DUB inhibitors directly into your lysis buffer. While PR-619 can be used, other options like N-ethylmaleimide (NEM) are commonly added to lysis buffers to instantly inactivate DUBs during the lysis process [36].
- Potential Cause 2: The PR-619 stock solution has degraded or was improperly prepared.
- Solution: Prepare a fresh stock solution in DMSO, ensure proper storage at -20°C, and avoid freeze-thaw cycles.
- Potential Cause 3: The ubiquitin signal is saturated or the gel/western transfer is not optimal for high-molecular-weight proteins.
- Solution: Optimize your gel electrophoresis and transfer conditions for high-MW proteins. Try loading less total protein to avoid signal saturation.
Problem 3: Difficulty in Interpreting Phenotype Due to Off-Target Effects
- Potential Cause: As a pan-DUB inhibitor, PR-619 simultaneously inhibits ~100 DUBs, making it challenging to attribute a phenotype to the inhibition of a specific DUB [20] [6].
- Solutions:
- Use PR-619 for initial, broad-target validation. Follow up with genetic approaches (e.g., siRNA, CRISPR knockout) targeting individual DUBs to confirm the phenotype.
- Employ more selective DUB inhibitors if available for your DUB of interest.
- Use activity-based probes (ABPs) like HA-Ub-VS to profile the DUB activity landscape in your cells and confirm which DUB families are being engaged by PR-619 [36] [35].
The Scientist's Toolkit: Essential Research Reagents
| Reagent / Tool | Primary Function | Key Application in DUB Research |
|---|---|---|
| PR-619 | Pan-deubiquitinase inhibitor | Induces global ubiquitin accumulation; used to study the effects of blocking deubiquitination [2] [37]. |
| HA-Ub-VS (Ubiquitin Probe) | Activity-based probe for DUBs | Covalently tags active DUBs in lysates; used to confirm DUB inhibition by PR-619 and profile active DUBs [36] [35]. |
| PYR-41 / TAK243 | Ubiquitin E1 Inhibitor | Blocks the ubiquitination cascade; used in combination studies to distinguish ubiquitination/depletion dynamics [2] [14]. |
| MG132 / Bortezomib | Proteasome Inhibitor | Blocks degradation of ubiquitinated proteins; used to compare effects of proteasome vs. DUB inhibition [14] [6]. |
| Chloroquine / Bafilomycin A1 | Autophagy Inhibitors | Blocks late-stage autophagy; used to determine the functional role of autophagy in PR-619's mechanism [2]. |
Experimental Protocol: Assessing DUB Inhibition in Cell Lysates
This protocol allows you to confirm that your PR-619 treatment is effectively inhibiting DUB activity during cell lysis and experimentation [36].
Step-by-Step Guide:
Cell Lysis with DUB Inhibition:
- Prepare two aliquots of cell pellet.
- Resuspend the first pellet in your standard lysis buffer.
- Resuspend the second pellet in lysis buffer supplemented with 20-50 µM PR-619 or 1 mM NEM.
- Incubate on ice for 10-30 minutes, then centrifuge to clear the lysate.
Protein Concentration Measurement:
- Perform a BCA or Bradford assay to determine the protein concentration of all lysates.
Western Blot Analysis for Ubiquitin:
- Load equal amounts of protein (e.g., 20-40 µg) from each lysate on an SDS-PAGE gel.
- Transfer to a PVDF membrane.
- Probe the membrane with an anti-ubiquitin antibody.
- Compare the ubiquitin smear between the standard lysis sample and the DUB-inhibited sample. A stronger, higher molecular weight smear in the DUB-inhibited sample indicates successful protection of ubiquitin conjugates during lysis.
Signaling Pathway Diagrams
PR-619-Induced Signaling Pathways
Workflow for DUB Activity Profiling
Cell lysis is a fundamental first step in molecular biology that involves breaking down cell membranes to release intracellular contents, including proteins, DNA, and RNA [39]. The selection and optimization of a lysis buffer are critical for successful downstream applications, particularly in specialized research such as studying ubiquitination pathways. For researchers investigating deubiquitination processes, proper lysis buffer formulation is essential to preserve post-translational modifications and prevent artificial loss of ubiquitin signals during sample preparation.
Lysis buffers are specially formulated solutions designed to disrupt cell membranes while maintaining the stability and integrity of intracellular components [39]. They typically contain a combination of detergents, salts, buffering agents, and enzyme inhibitors tailored to specific cell types and compatible with various downstream applications [40]. The optimization of these components—their concentrations, incubation parameters, and compatibility with inhibitors—forms the foundation of reliable and reproducible experimental outcomes in deubiquitination research.
Core Components of Lysis Buffers
Buffering Agents and Salts
The buffering system in a lysis buffer maintains a stable pH environment, which is crucial for protein stability and activity. The choice of buffer depends on the desired pH range and compatibility with the protein of interest and downstream applications [40].
Common Buffering Agents and Their Properties:
| Buffer | pH Range | Key Characteristics | Compatibility Notes |
|---|---|---|---|
| Tris-HCl | 7.0 - 9.0 | Widely used, cost-effective | May interfere with some downstream assays |
| HEPES-NaOH | 7.2 - 8.2 | Good buffering capacity in physiological range | Better for enzyme activity studies |
| Sodium phosphate | 5.8 - 8.0 | Broad range | Compatibility varies |
Salts are incorporated into lysis buffers to regulate ionic strength and osmolarity. Commonly used salts include NaCl and KCl, typically in concentrations between 50-150 mM [40]. Optimization studies have demonstrated that adjusting NaCl concentration can significantly impact protein yield during solubilization [41]. Additionally, metal chelators like EDTA are often included to bind metal ions that could activate proteases [42].
Detergents and Solubilizing Agents
Detergents are amphipathic molecules that disrupt lipid bilaries and solubilize membrane proteins. They are categorized based on their hydrophilic head groups into ionic (anionic, cationic), nonionic, and zwitterionic classes, each with different properties and applications [40].
Detergent Classification and Properties:
| Detergent Type | Examples | Denaturing Properties | Recommended Use Cases |
|---|---|---|---|
| Nonionic | Triton X-100, NP-40 | Non-denaturing | Preserving protein-protein interactions; immunoprecipitation |
| Anionic | SDS | Denaturing | Solubilizing difficult proteins; complete denaturation |
| Zwitterionic | CHAPS | Non-denaturing | Membrane protein studies; maintaining protein activity |
The concentration of detergents is critical for efficient lysis. For nonionic detergents like Triton X-100 or NP-40, concentrations around 1% are typically effective [43] [42]. Stronger denaturing detergents like SDS are used at varying concentrations (0.1-1%) depending on the required stringency [41] [44]. Research has shown that optimizing SDS concentration in lysis buffers significantly enhances protein solubilization from precipitated pellets [41].
Enzyme Inhibitors and Additives
Protease and phosphatase inhibitors are essential additives that prevent protein degradation and preserve post-translational modifications during lysis. For deubiquitination research, including deubiquitinase (DUB) inhibitors is crucial to prevent artificial loss of ubiquitin signals.
Essential Inhibitors for Lysis Buffers:
| Inhibitor Type | Target Enzymes | Recommended Concentration | Stability Considerations |
|---|---|---|---|
| PMSF | Serine proteases | 1 mM | Short half-life in aqueous solution; add fresh |
| EDTA | Metalloproteases | 1-5 mM | Stable in solution |
| Sodium orthovanadate | Phosphatases | 0.2-2 mM | Requires activation |
| Protease inhibitor cocktails | Broad-spectrum proteases | As manufacturer recommends | Typically added fresh |
| DUB inhibitors | Deubiquitinases | Variable by product | Specific to research needs |
Inhibitors should be added fresh to lysis buffer immediately before use, as they can degrade upon storage—even at -20°C, particularly in frost-free freezers [43]. Additional additives like glycerol (5-10%) can help stabilize protein structures, while reducing agents like DTT (1-5 mM) or β-mercaptoethanol (0.1-1%) break disulfide bonds for complete denaturation [45].
Optimization Parameters for Lysis Buffers
Concentration Optimization
Systematic optimization of lysis buffer components significantly enhances protein yield and quality. Research demonstrates that tailored concentrations of SDS, NaCl, and EDTA in lysis buffers can dramatically improve protein solubilization efficiency, particularly for difficult samples like TRIzol-precipitated protein pellets [41].
Optimized Concentration Ranges for Lysis Buffer Components:
| Component | Standard Concentration | Optimized Range | Impact on Protein Yield |
|---|---|---|---|
| SDS | 0.1% (in RIPA) | 0.1-1% | Significant improvement in solubilization |
| NaCl | 150 mM | 50-200 mM | Moderate to significant improvement |
| EDTA | 1-2 mM | 1-5 mM | Moderate improvement |
| Tris-HCl | 10-50 mM | 10-50 mM | Minimal individual impact |
| Nonionic detergents | 0.5-1% | 0.5-2% | Concentration-dependent efficiency |
Studies have shown that adjusting these components in a standard lysis buffer increased protein yield during solubilization and was more effective at directly homogenizing brain tissue than standard RIPA buffer [41]. The optimized buffer composition also effectively represented different neural cell types and protein classes in the solubilized samples [41].
Incubation Timing and Temperature
Proper incubation parameters are essential for efficient lysis while maintaining protein integrity. Standard protocols typically recommend incubating samples in lysis buffer on ice for 15-30 minutes [33] [44]. However, research indicates that effective incubation parameters for both total protein yield and analysis of post-translational modifications can be remarkably flexible across various temperatures and durations [41].
Incubation Parameter Optimization:
| Temperature | Recommended Duration | Application Context |
|---|---|---|
| 4°C (on ice) | 15-30 minutes | Standard protocol; preserves protein interactions |
| Room temperature | 5-15 minutes | Rapid processing; compatible with some inhibitors |
| 37°C | 5-10 minutes | Enhanced efficiency for some cell types |
| 50°C | Varies (with validation) | Specialized applications |
| 90-95°C | 10-20 minutes | Complete denaturation for SDS lysis |
For most applications, keeping samples on ice during lysis is recommended to minimize protein degradation [44] [45]. When using hot SDS lysis buffer, samples are typically boiled at 90-95°C for 10-20 minutes [44]. It's important to note that optimal incubation conditions may require empirical determination for specific sample types and research objectives.
Lysis Buffer Compatibility and Selection
Compatibility with Downstream Applications
The choice of lysis buffer must align with intended downstream applications, as buffer components can significantly impact subsequent experimental steps.
Lysis Buffer Compatibility with Common Applications:
| Application | Recommended Buffer Type | Compatibility Considerations |
|---|---|---|
| Western blotting | RIPA, SDS, NP-40 | Most buffers compatible; consider denaturation needs |
| Immunoprecipitation/Co-IP | NP-40, IP Lysis Buffer | Avoid strong denaturants like SDS |
| Protein quantification | Compatible with assay | SDS may interfere with some assays |
| Mass spectrometry | Urea-based, RIPA | Detergent removal may be necessary |
| Enzyme activity assays | Mild nonionic detergents | Avoid denaturing conditions |
RIPA buffer is a popular choice for general protein extraction and western blotting as it effectively extracts membrane, cytoplasmic, and nuclear proteins [39] [42]. However, for techniques like immunoprecipitation that require preserved protein-protein interactions, milder buffers such as NP-40 or specialized IP lysis buffers are more appropriate [39] [42].
Sample-Specific Buffer Selection
Different sample types require tailored lysis approaches for optimal protein extraction.
Recommended Lysis Buffers by Sample Type:
| Sample Type | Recommended Buffer | Alternative Options | Special Considerations |
|---|---|---|---|
| Adherent mammalian cells | RIPA, NP-40 | M-PER, IP Lysis Buffer | Mechanical scraping may be needed |
| Suspension mammalian cells | RIPA, NP-40 | M-PER, Cell Lysis Buffer | Centrifugation steps required |
| Brain tissue | Optimized lysis buffer, N-PER | RIPA, T-PER | High lipid content; homogenization essential |
| Other tissues (liver, heart) | T-PER, RIPA | SDS buffer | Mechanical homogenization required |
| Bacterial cells | B-PER, BugBuster | Urea-based | Cell wall requires stronger disruption |
| Plant cells | specialized plant buffers | Urea-based, SDS | Rigid cell wall requires vigorous methods |
For mammalian tissues, specialized reagents like T-PER (for general tissues) and N-PER (for neuronal tissues) have been developed to optimize protein extraction efficiency [39]. Research demonstrates that optimized lysis buffer formulations can be particularly beneficial for difficult-to-attain samples, such as specific sorted cell populations [41].
Troubleshooting Common Lysis Buffer Issues
Frequently Asked Questions
Why is my protein yield low?
- Cause: Inefficient lysis, improper detergent concentration, or protein degradation [43].
- Solution: Verify detergent concentration (typically 1% for nonionic detergents) [43]. Ensure fresh protease inhibitors are added immediately before use [43]. For difficult samples, consider using a stronger denaturing buffer with SDS or urea [41] [42]. Pre-cool all equipment and work quickly on ice to minimize degradation [44] [45].
How can I prevent protein degradation during lysis?
- Cause: Inadequate inhibition of endogenous proteases, including deubiquitinases.
- Solution: Always add fresh protease inhibitors immediately before lysis [43]. Include specific DUB inhibitors for ubiquitination studies. Keep samples on ice throughout the process and use pre-chilled buffers [44] [45]. For particularly sensitive samples, consider flash-freezing in liquid nitrogen before lysis [44].
My lysate is viscous—what should I do?
- Cause: Release of genomic DNA during lysis.
- Solution: Briefly sonicate samples (3-5 pulses of 2-3 seconds with intervals) [33] [44]. Alternatively, add DNase I (without Mg²⁺ to prevent activation of magnesium-dependent enzymes) or use lysis buffers containing a universal nuclease [17].
Why is my background high in western blots?
- Cause: Non-specific binding or incomplete lysis.
- Solution: Ensure proper centrifugation after lysis (10,000-15,000 × g for 10-20 minutes at 4°C) to remove insoluble debris [44] [45]. Optimize detergent concentration and consider switching to a more specific buffer formulation for your target protein [42].
How do I maintain ubiquitin signals during lysis?
- Cause: Activity of endogenous deubiquitinases (DUBs).
- Solution: Include specific DUB inhibitors in your lysis buffer. Work rapidly at 4°C to minimize enzyme activity. Consider using stronger denaturing conditions (e.g., SDS buffer with boiling) to instantly inactivate DUBs [44]. Validate your protocol with known ubiquitinated standards.
Advanced Troubleshooting Guide
| Problem | Potential Causes | Recommended Solutions | Prevention Tips |
|---|---|---|---|
| Insoluble proteins | Denatured aggregates, inclusion bodies | Use urea (2-8M) or guanidine-HCl for solubilization | Optimize expression conditions; test multiple buffers |
| Incomplete lysis | Insufficient detergent, wrong buffer type | Increase detergent concentration; switch buffer type | Validate buffer against cell type; include mechanical disruption |
| Enzyme activity loss | Denaturing conditions | Switch to mild nonionic detergents (NP-40, Triton X-100) | Use non-denaturing buffers; avoid SDS and deoxycholate |
| Interference with quantification | Detergent incompatibility | Use detergent-compatible assays (BCA) | Dilute samples; include buffer controls in standard curve |
| Inconsistent results | Variable inhibitor efficacy | Prepare fresh inhibitor cocktails; standardize protocols | Aliquot inhibitors; establish standardized workflows |
Essential Research Reagent Solutions
Key Research Reagents for Lysis Buffer Preparation:
| Reagent Category | Specific Examples | Function | Application Notes |
|---|---|---|---|
| Protease inhibitors | PMSF, AEBSF, protease inhibitor cocktails | Inhibit serine, cysteine, metallo proteases | Add fresh; consider specific protease profiles |
| Phosphatase inhibitors | Sodium orthovanadate, sodium fluoride | Preserve phosphorylation states | Essential for phosphoprotein studies |
| DUB inhibitors | PR-619, N-ethylmaleimide | Prevent deubiquitination | Crucial for ubiquitination research |
| Detergents | NP-40, Triton X-100, SDS, CHAPS | Membrane disruption and protein solubilization | Select based on denaturation needs |
| Commercial lysis buffers | RIPA, NP-40, M-PER, T-PER | Optimized formulations | Save preparation time; ensure consistency |
| Nuclease reagents | DNase I, RNase A, universal nuclease | Reduce viscosity from nucleic acids | Improve sample handling and loading |
Experimental Protocols
Standard Protocol for Cell Lysis with Optimized Buffer
Materials:
- Cultured cells (adherent or suspension)
- Ice-cold PBS
- Optimized lysis buffer (see formulation below)
- Fresh protease/phosphatase/DUB inhibitors
- Cell scraper (for adherent cells)
- Refrigerated centrifuge
- Sonicator
Optimized Lysis Buffer Formulation:
- 50 mM Tris-HCl, pH 7.4
- 150 mM NaCl
- 1% NP-40 or Triton X-100
- 0.5% sodium deoxycholate
- 0.1% SDS
- 1 mM EDTA
- Add fresh inhibitors immediately before use: 1 mM PMSF, protease inhibitor cocktail, phosphatase inhibitors, and DUB inhibitors as needed [41] [42]
Procedure:
- Prepare cells: For adherent cells, wash with ice-cold PBS and scrape into cold PBS. For suspension cells, pellet by centrifugation at 300 × g for 5 minutes at 4°C and wash with PBS [33] [45].
- Pellet cells: Centrifuge at 1000 × g for 5 minutes at 4°C to pellet cells [42].
- Add lysis buffer: Resuspend cell pellet in optimized lysis buffer (100 μL per 10⁶ cells) [42].
- Incubate: Place on ice for 30 minutes with occasional vortexing [42] [45].
- Sonicate: Sonicate briefly (3-5 pulses of 2-3 seconds with 10-second intervals on ice) to reduce viscosity and ensure complete lysis [44].
- Clarify: Centrifuge at 10,000-12,000 × g for 20 minutes at 4°C [42] [45].
- Collect supernatant: Transfer clarified lysate to a new tube without disturbing the pellet [45].
- Quantify and store: Determine protein concentration using a compatible assay and store at -80°C for long-term storage [45].
Protocol for Tissue Lysis with Deubiquitination Protection
Materials:
- Tissue samples (fresh or frozen)
- Liquid nitrogen
- Mortar and pestle or tissue homogenizer
- Optimized lysis buffer with DUB inhibitors
- Ice-cold PBS
Procedure:
- Prepare tissue: Dissect tissue of interest and wash with ice-cold PBS to remove blood if necessary [45].
- Flash-freeze (optional): For frozen tissues, snap-freeze in liquid nitrogen [44] [45].
- Homogenize: Grind frozen tissue to a powder in liquid nitrogen using a mortar and pestle, or homogenize fresh tissue directly in lysis buffer using an electric homogenizer [44] [45].
- Add lysis buffer: Add 500 μL of optimized lysis buffer with fresh DUB inhibitors per 10 mg of tissue [42].
- Incubate: Maintain constant agitation for 2 hours at 4°C [45].
- Clarify: Centrifuge at 12,000 × g for 20 minutes at 4°C [45].
- Collect supernatant: Transfer clarified lysate to a new tube, avoiding the lipid layer and pellet [45].
- Quantify: Determine protein concentration and adjust as needed for downstream applications [45].
Figure 1: Experimental Workflow for Optimized Protein Extraction*
Figure 2: DUB Inhibitor Protection of Ubiquitin Signals*
Integrating DUB Inhibitors with Standard Protease and Phosphatase Cocktails
Preventing undesired protein deubiquitination during cell lysis is a critical step in researching the ubiquitin-proteasome system (UPS). Deubiquitinating enzymes (DUBs), a family of approximately 100 proteases in human cells, cleave ubiquitin from protein substrates, thereby regulating protein stability, localization, and activity [46]. When a cell is lysed for experimentation, the controlled cellular environment is disrupted, allowing DUBs to artificially remove ubiquitin signals from proteins of interest. This can lead to the loss of crucial post-translational modification data and inaccurate conclusions about protein regulation, particularly in disease contexts like cancer and neurodegeneration where DUBs are heavily implicated [29] [46]. The strategic integration of DUB inhibitors into standard lysis cocktails, which already contain protease and phosphatase inhibitors, is therefore essential for preserving the native ubiquitination state of proteins and ensuring experimental integrity.
Frequently Asked Questions (FAQs)
1. Why is it necessary to include DUB inhibitors in my lysis buffer? During cell lysis, the compartmentalization that naturally regulates DUB activity is lost. This can lead to the rapid and artificial removal of ubiquitin chains from your protein targets before you can analyze them. DUB inhibitors covalently modify the active site of functional DUBs, irreversibly blocking their activity and "freezing" the ubiquitination state of proteins at the moment of lysis [18]. This is especially critical for studying proteins whose stability is regulated by the UPS or for detecting ubiquitin signals in pathways where DUBs are therapeutic targets [29] [46].
2. Can I simply add a DUB inhibitor to my commercial protease/phosphatase cocktail? While physically possible, the effectiveness depends on the compatibility of the buffers and the specific inhibitors. Commercial cocktails may have optimized pH and salt concentrations for serine, cysteine, and metallo-protease inhibition, which might not be ideal for all DUB inhibitors. It is recommended to either:
- Verify Compatibility: Consult the datasheets for pH and buffer requirements.
- Use a Pre-formulated Cocktail: Some manufacturers now offer lysis buffers that include DUB inhibitors.
- Reconstitute Separately: Reconstitute your DUB inhibitor in the recommended solvent (often DMSO) and add it to your lysis buffer immediately before use, alongside your standard cocktail.
3. What are the most common types of DUB inhibitors used during lysis? Common inhibitors target the active site cysteine present in several DUB subfamilies (e.g., USP, UCH, OTU). These include:
- Broad-Spectrum Inhibitors: PR-619 and HBX 41-1082 are often used for initial experiments to broadly preserve ubiquitination, as they target a wide range of DUBs [29].
- Selective Inhibitors: For studies focused on a specific DUB, more selective compounds are available (e.g., inhibitors for USP7, USP30) [29] [46]. The choice between broad-spectrum and selective inhibitors depends on whether you aim to preserve all ubiquitin signals or interrogate a specific DUB-substrate relationship.
4. My ubiquitination signal is still weak after adding inhibitors. What could be wrong? Several factors could be at play:
- Inhibitor Concentration: The inhibitor may be too dilute. Increase the concentration within the manufacturer's recommended range.
- Incorrect Lysis Buffer: Ensure your lysis buffer is compatible. Harsh denaturing conditions (e.g., containing SDS) may be necessary for some applications to fully inactivate enzymes immediately [47].
- Insufficiently Rapid Processing: Even with inhibitors, keeping samples on ice and processing them quickly is vital.
- Deubiquitination After Lysis: Deubiquitination can occur during subsequent steps like immunoprecipitation if inhibitors are not also present in all buffers [47].
Troubleshooting Guide
Table 1: Common Problems and Solutions when Using DUB Inhibitors
| Problem | Potential Cause | Recommended Solution |
|---|---|---|
| High background or smeared bands in western blot for ubiquitin. | Incomplete inhibition of DUBs leading to partial, non-specific deubiquitination. | Increase the concentration of the DUB inhibitor. Switch to a more denaturing lysis buffer (e.g., 2% SDS) and boil samples immediately [47]. |
| No detectable ubiquitin signal. | Overwhelming DUB activity not being controlled; protein not ubiquitinated; inhibitor is degraded or inactive. | Verify inhibitor is fresh and stored correctly. Include a positive control known to be ubiquitinated. Use a combination of broad-spectrum DUB inhibitors. |
| Cell lysis is inefficient. | Lysis buffer composition is affected by the DUB inhibitor solvent (e.g., high DMSO concentration). | Reduce the volume of solvent added; ensure the final DMSO concentration is ≤1%. Consider using a different inhibitor that is water-soluble. |
| Inconsistent results between experiments. | Inconsistent addition of the DUB inhibitor to the lysis buffer; variation in sample handling time. | Create a large, single-use batch of complete lysis buffer with all inhibitors, then aliquot and freeze it. Standardize the time between lysis and boiling/analysis. |
The Scientist's Toolkit: Essential Reagents
Table 2: Key Research Reagent Solutions for DUB Inhibition
| Reagent | Function in Experiment | Key Considerations |
|---|---|---|
| Broad-Spectrum DUB Inhibitors (e.g., PR-619) | Covalently modifies active site cysteine of many DUBs to preserve global ubiquitination states during lysis [29]. | Ideal for initial, discovery-phase experiments. May complicate identifying which specific DUB is acting on a substrate. |
| Selective DUB Inhibitors (e.g., for USP7, USP30) | Inhibits a specific DUB to study its particular biological function and substrate profile [29] [46]. | Essential for target validation and mechanistic studies. Selectivity should be confirmed in your cellular model. |
| Activity-Based Probes (ABPs) (e.g., HA-Ub-VS) | Ubiquitin-based probes that covalently label active DUBs in lysates, allowing for detection and quantification of functional DUB levels on a western blot [18] [29]. | Useful for verifying that your DUB inhibitor is effectively blocking active sites and for profiling DUB activity in different cell lines. |
| Denaturing Lysis Buffer (e.g., 2% SDS) | Rapidly denatures all enzymes, including DUBs, at the moment of lysis, providing the most robust preservation of post-translational modifications [47]. | Required for certain ubiquitination assays. Compatible with downstream applications like immunoblotting but may not be suitable for native immunoprecipitation without dilution. |
| Protease & Phosphatase Inhibitor Cocktails | Standard cocktails inhibit a range of serine, cysteine, and metallo-proteases, as well as phosphatases, to prevent general protein degradation and modification loss. | DUB inhibitors are a critical supplement to these standard cocktails, not a replacement. They should be used concurrently. |
Detailed Experimental Protocols
Protocol 1: Cell Lysis with Integrated DUB Inhibition for Western Blotting
This protocol is designed for preparing cell lysates specifically for the analysis of protein ubiquitination by western blot.
Materials:
- Lysis Buffer (e.g., RIPA or a 2% SDS buffer for full denaturation: 2% SDS, 150 mM NaCl, 10 mM Tris-HCl, pH 8.0) [47]
- Protease Inhibitor Cocktail (without DUB inhibitors)
- Phosphatase Inhibitor Cocktail
- DUB Inhibitor (e.g., 1-10 mM PR-619 or a selective inhibitor in DMSO)
- Dithiothreitol (DTT)
- PBS (Phosphate Buffered Saline)
Method:
- Prepare Complete Lysis Buffer: Add protease inhibitor cocktail, phosphatase inhibitor cocktail, and the DUB inhibitor to the lysis buffer immediately before use. Optionally, include 1-2 mM DTT to maintain a reducing environment [18].
- Harvest and Wash Cells: Culture and harvest your cells according to your standard method. Wash the cell pellet with ice-cold PBS to remove residual media proteins.
- Lyse Cells: Add the complete, ice-cold lysis buffer directly to the cell pellet (e.g., 100 µl for a 6 cm dish) [47]. Vortex briefly to mix.
- Immediately Denature: For the most robust preservation, transfer the lysate to a heat block or water bath and boil at 95-100°C for 5-10 minutes immediately after adding the buffer [47].
- Sonicate and Clarify: Sonicate the boiled lysate to shear genomic DNA and reduce viscosity. Centrifuge the lysate at high speed (e.g., 20,000 x g for 10-15 minutes) to pellet insoluble debris.
- Collect and Quantify: Transfer the clarified supernatant to a new tube. Quantify the protein concentration using a compatible assay (e.g., BCA assay) before proceeding to SDS-PAGE and western blotting.
Protocol 2: Validating DUB Inhibition with Activity-Based Probes
This method allows you to confirm that the DUB inhibitors in your lysis buffer are effectively blocking DUB activity.
Materials:
- HA-Ub-Vinyl Sulfone (HA-Ub-VS) or similar activity-based probe [18]
- Control and treated cell lysates
- Laemmli's sample buffer
- Standard Western Blot equipment and reagents
- Anti-HA antibody
Method:
- Prepare Lysates: Generate two aliquots of cell lysate. Lyse one with your standard cocktail (Control), and the other with the standard cocktail plus your DUB inhibitor (Treated). Do not boil these lysates yet.
- Incubate with Probe: Add the HA-Ub-VS probe (e.g., 50 nM final concentration) to both lysates and incubate at 37°C for 1 hour [18]. The probe will covalently tag any actively functioning DUBs.
- Stop Reaction: Add Laemmli's sample buffer to the lysates and boil for 5 minutes at 95°C.
- Analyze by Western Blot: Run the samples on an SDS-PAGE gel, transfer to a membrane, and blot with an anti-HA antibody.
- Interpret Results: Successful DUB inhibition in the Treated sample will be evidenced by a significant reduction or complete absence of labeled DUB bands compared to the Control lysate, confirming the efficacy of your inhibitor cocktail [18].
Workflow and Pathway Diagrams
Experimental Workflow for Preserving Ubiquitination
DUB Function and Inhibitor Mechanism
The study of protein ubiquitination is crucial for understanding diverse cellular processes, ranging from protein degradation to signal transduction. For researchers investigating ubiquitin-dependent pathways, preventing artifactual deubiquitination during cell lysis is a fundamental experimental concern. Deubiquitinases (DUBs) remain highly active under standard lysis conditions and can rapidly remove ubiquitin modifications, leading to inaccurate experimental results. This technical guide provides optimized protocols for co-immunoprecipitation (Co-IP) and ubiquitin pulldown experiments, with emphasis on preserving the native ubiquitin landscape through strategic DUB inhibition. The following sections address common challenges and provide workflow-specific solutions for maintaining ubiquitin modifications throughout experimental procedures.
Frequently Asked Questions (FAQs) and Troubleshooting Guides
FAQ: DUB Inhibition and Lysis Conditions
Q1: Why is it necessary to include DUB inhibitors during cell lysis?
DUBs are cysteine proteases that remain enzymatically active under standard cell lysis conditions. Without inhibition, they can rapidly remove ubiquitin signals from your protein of interest, resulting in false-negative outcomes and compromised data. Research demonstrates that many DUBs are sensitive to oxidative inhibition, but this natural regulatory mechanism is insufficient for experimental preservation of ubiquitination states [5].
Q2: What type of DUB inhibitor should I use?
The choice of DUB inhibitor depends on your experimental goals. Broad-spectrum DUB inhibitors like PR-619 provide robust protection against multiple DUB families and are ideal for initial experiments. For more specific targeting, selective inhibitors against particular DUB classes (USP, UCH, etc.) may be preferable. PR-619 has been shown to induce accumulation of ubiquitylated proteins and trigger endoplasmic reticulum stress in experimental models [2].
Q3: How should DUB inhibitors be prepared and stored?
Most DUB inhibitors are reconstituted in DMSO and stored as small aliquots at -20°C or -80°C. Avoid repeated freeze-thaw cycles. Always add inhibitors to lysis buffer immediately before use, as some components may degrade over time in aqueous solution.
Q4: Can I combine DUB inhibitors with other protease inhibitors?
Yes, DUB inhibitors should be used in conjunction with standard protease inhibitor cocktails (targeting serine, cysteine, aspartic, and metalloproteases) and phosphatase inhibitors to comprehensively preserve post-translational modifications.
Troubleshooting Common Experimental Issues
Problem: Low yield of ubiquitinated proteins in pulldown experiments
Potential Causes and Solutions:
- Insufficient DUB inhibition: Increase concentration of DUB inhibitors in lysis buffer and confirm inhibitor activity.
- Improper lysis conditions: Ensure complete cell disruption while maintaining non-denaturing conditions appropriate for protein complex preservation.
- Antibody issues: Validate antibody specificity for ubiquitinated forms of your target protein.
- Epitope masking: Consider using tags (e.g., Strep-tag, FLAG) instead of antibodies for purification when possible [48].
Problem: High background or non-specific binding
Potential Causes and Solutions:
- Insufficient washing: Increase wash stringency by adding non-ionic detergents (0.01-0.1% Tween-20 or Triton X-100) to wash buffers [49].
- Antibody concentration too high: Titrate antibody to optimal concentration to reduce non-specific binding.
- Incomplete blocking: Ensure blocking agents are included in wash buffers when necessary.
- Consider pre-clearing: Perform a pre-clearing step with bare beads to remove proteins that bind non-specifically.
Problem: Antibody heavy/light chains interfering with western blot analysis
Potential Causes and Solutions:
- Crosslink antibodies to beads: Covalently attach antibodies to resin using commercial crosslinking kits [49].
- Use different species for blotting: Use secondary antibodies from different species than your IP antibody.
- Specialized detection reagents: Implement Clean-Blot IP Detection Reagent which detects only native antibodies without recognizing denatured heavy/light chains [49].
Problem: Protein complex disruption during elution
Potential Causes and Solutions:
- Overly harsh elution conditions: Test gentler elution buffers such as high-salt, neutral pH elution buffer [49].
- Avoid reducing agents: Exclude DTT or β-mercaptoethanol from elution buffers unless necessary, as they can dissociate antibody chains [49].
Research Reagent Solutions for Ubiquitin Studies
Table: Essential Reagents for Ubiquitin Pulldown and Co-IP Experiments
| Reagent Type | Specific Examples | Function/Application | Key Considerations |
|---|---|---|---|
| DUB Inhibitors | PR-619 | Pan-DUB inhibitor; induces ub-protein accumulation [2] | Use at 10-50 μM in lysis buffers; prepare fresh in DMSO |
| Affinity Beads | Streptavidin magnetic beads | Bind biotin-tagged proteins/peptides in pulldown assays [50] | Use 2 μL beads per mL lysate; wash before use |
| Protease Inhibitors | EDTA-free protease inhibitor cocktails | Prevent general protein degradation | Compatible with metal-dependent processes |
| Lysis Buffers | Modified RIPA, NP-40 based | Extract proteins while preserving complexes | Include 0.05% NP-40 to facilitate pull-down reactions [50] |
| Ubiquitin Sources | Human synthetic ubiquitin | Ubiquitin moiety donor for in vitro assays [50] | Use at 1 μg per pull-down reaction |
| Tagging Systems | Strep-tag, FLAG, HIS | Enable affinity purification without antibodies [48] | Strep-tag offers high specificity and gentle elution |
| Crosslinkers | DSS, BS3 | Covalently attach antibodies to beads | Prevent antibody co-elution [49] |
Table: Critical Parameters for Ubiquitin Workflows
| Experimental Parameter | Recommended Range | Optimal Value | Notes |
|---|---|---|---|
| DUB Inhibitor Concentration | 10-50 μM | 20 μM | PR-619 effective in this range [2] |
| Lysis Buffer pH | 7.0-8.0 | 7.4 | Maintain physiological conditions |
| Incubation Time with Antibody/Beads | 2 hours to overnight | 4 hours | Balance between yield and specificity |
| Wash Stringency (Detergent) | 0.01-0.1% | 0.05% | Tween-20 or Triton X-100 [49] |
| Number of Washes | 3-5 | 4 | Fewer washes increase background; more may disrupt complexes |
| Elution Buffer pH | 2.0-3.0 or neutral | Based on application | Low pH denatures; neutral pH preserves complexes [49] |
| Post-lysis Processing Time | <30 minutes | Immediately | Minimize DUB activity before inhibition |
Advanced Methodologies
PTM-Enhanced Pull-Down Assay
The Post-Translational Modification-enhanced (PTMe) pull-down method represents a significant advancement for studying ubiquitin E-ligase complexes and phospho-degron interactions. This integrated approach combines kinase and ubiquitination assays within a single pull-down step using cell extracts as a source of enzymatically active modification proteins [50].
Key features of PTMe pull-down:
- Suitable for studying UPS-regulated cytosolic and nuclear proteins
- Requires biotin-tagged recombinant version of target protein/domain
- Enables analysis of endogenous ubiquitin E-ligase recruitment
- Allows simultaneous testing of various phosphorylation/ubiquitination conditions
Protocol Summary:
- Prepare biotin-tagged peptide containing putative phospho-degron motif
- Couple with streptavidin magnetic beads
- Incubate with cell extract containing:
- ATP (4 mM as phosphate donor)
- DTT (4 mM as reducing agent)
- Ubiquitin (1 μg/reaction as ubiquitin donor)
- MnCl₂ and MgCl₂ (2 mM each as kinase cofactors)
- Wash and elute bound complexes
- Analyze by western blot or mass spectrometry [50]
Tandem Affinity Purification (TAP) for Complex Isolation
Tandem Affinity Purification provides enhanced specificity for isolating protein complexes under near-physiological conditions [48].
Workflow Overview:
- Genetically fuse TAP tag to protein of interest
- Express in appropriate cellular system
- Perform sequential purification using two distinct affinity tags
- Elute and analyze purified complexes
Advantages over single-step purification:
- Increased specificity through dual purification steps
- Enhanced purity of isolated complexes
- Versatility across physiological conditions
- Compatibility with downstream proteomic analysis [48]
Visual Experimental Workflows
Ubiquitin Pull-Down Workflow with DUB Inhibition
Integrated PTM-Enhanced Pull-Down Methodology
Maintaining ubiquitin modifications during cell lysis and protein purification requires careful attention to DUB inhibition strategies. The protocols and troubleshooting guides presented here provide a foundation for reliable co-IP and ubiquitin pulldown experiments. By implementing these workflow-specific recommendations—particularly the consistent use of DUB inhibitors during initial cell lysis—researchers can significantly improve the accuracy of their ubiquitination studies and obtain more biologically relevant data for both basic research and drug development applications.
Addressing Technical Challenges in DUB Inhibition and Artifact Prevention
{ article }
Selectivity vs. Efficacy: Balancing Comprehensive Inhibition Against Experimental Goals
Core Concepts & FAQ
Frequently Asked Questions
Why is it crucial to inhibit deubiquitinases (DUBs) during cell lysis? During cell lysis, the compartmentalization that separates DUBs from their substrates is lost. This allows DUBs to rapidly remove ubiquitin signals from proteins before they can be analyzed. Using DUB inhibitors in your lysis buffer preserves the native ubiquitin landscape by preventing this post-lysis deubiquitination, ensuring that your experimental results reflect the true cellular state [14].
How do I choose between a pan-DUB inhibitor and a selective inhibitor for my lysis buffer? The choice depends entirely on your experimental goal. Use a broad-spectrum, pan-DUB inhibitor (e.g., PR-619) when you aim to preserve the global ubiquitinome for an unbiased analysis, such as in mass spectrometry-based proteomic studies. Conversely, use a selective DUB inhibitor when your research focuses on the specific function of a particular DUB or when you are validating a suspected DUB target in a biological pathway [29] [51].
My ubiquitin signal is still weak after adding a DUB inhibitor. What could be wrong? This is a common issue with several potential causes:
- Incorrect Inhibitor Concentration: The inhibitor may be too dilute. Re-check the recommended working concentration and ensure your stock solution is fresh.
- Improper Lysis Buffer Composition: The detergent in your buffer might be incompatible or insufficient for complete cell disruption, leading to low protein yield and signal. Ensure you are using a recommended, effective lysis buffer [17].
- Oxidation of the DUB Inhibitor: Many DUB inhibitors are cysteine protease inhibitors whose active site cysteine is sensitive to oxidation. Always include reducing agents like Dithiothreitol (DTT) in your lysis buffer to maintain inhibitor activity [5].
- Insufficient Lysis: Low protein concentration can simply mean the cells are not fully lysed. Increase lysis buffer volume relative to cell pellet size, extend incubation time, or use more vigorous agitation [52].
The Scientist's Toolkit: Research Reagent Solutions
The following table details key reagents essential for successful experiments involving DUB inhibition.
Table 1: Essential Reagents for DUB Inhibition during Cell Lysis
| Item | Function & Rationale | Example |
|---|---|---|
| Pan-DUB Inhibitor | Broadly inhibits a wide range of cysteine-dependent DUBs to preserve the global ubiquitinome. Ideal for discovery-phase experiments. | PR-619 [2] [14] |
| Selective DUB Inhibitors | Targets a specific DUB (e.g., USP14) to investigate its unique biological function or validate it as a drug target. | VLX1570 (targets USP14) [53] |
| Proteasome Inhibitor | Often used in conjunction with DUB inhibitors to fully block protein degradation and stabilize ubiquitinated substrates. | MG132, Bortezomib, Carfilzomib [14] |
| Reducing Agent | Critical for maintaining the activity of cysteine-dependent DUB inhibitors by preventing oxidation of the catalytic cysteine. | Dithiothreitol (DTT) [5] |
| Protease Inhibitor Cocktail | Prevents general protein degradation by serine, cysteine, and metallo proteases during and after lysis. | cOmplete Lysis-M [52] |
| Nuclease | Reduces lysate viscosity by digesting genomic DNA and RNA, which is released upon cell disruption. This is particularly important for efficient pipetting and subsequent analyses. | Benzonase [54] |
| Metal Chelator | Inhibits metalloproteases that can degrade proteins. Note: Should be omitted if your protein purification strategy involves metal affinity chromatography (e.g., Ni-NTA for His-tagged proteins). | EDTA [54] |
Troubleshooting DUB Inhibition Experiments
This section provides a structured guide to diagnosing and resolving common problems.
Table 2: Troubleshooting Guide for DUB Inhibition Experiments
| Problem | Potential Cause | Recommended Solution |
|---|---|---|
| Low Protein Yield | Inefficient cell lysis due to insufficient lysis buffer volume or weak detergent. | Increase lysis buffer volume to 2-5 times the cell pellet volume. Use a more effective detergent and ensure vigorous mixing during incubation [52] [54]. |
| High Background in Ubiquitin Blots | Incomplete inhibition of DUBs leading to non-specific deubiquitination and smear. | Confirm inhibitor is fresh and used at correct concentration. Add a reducing agent (DTT) to lysis buffer. Consider using a combination of pan- and proteasome-specific inhibitors [5] [14]. |
| Loss of Protein Solubility/Aggregation | Lysis buffer is not optimal for your protein of interest; hydrophobic proteins aggregating. | Adjust lysis buffer composition (salt, detergent). Avoid freezing/thawing lysates. Clarify lysate by high-speed centrifugation immediately after lysis [17] [54]. |
| Inconsistent Results Between Experiments | Oxidation of DUB inhibitors; slight variations in lysis buffer pH or composition. | Always prepare lysis buffer fresh with reducing agents. Standardize buffer recipes and cell counting methods. Use validated inhibitor stock solutions [5]. |
Experimental Protocols & Data
Standardized Lysis Protocol with DUB Inhibition
This protocol is designed for mammalian cells and ensures preservation of ubiquitination states.
Preparation: Pre-chill centrifuge and all equipment to 4°C. Prepare Fresh Lysis Buffer containing:
- 25-50 mM Tris-HCl, pH 8.0
- 150-300 mM NaCl
- 1-2% recommended detergent (e.g., NP-40)
- 10% Glycerol
- 1-10 µM DUB Inhibitor (e.g., PR-619)
- 1-5 mM DTT
- 1x Protease Inhibitor Cocktail (without EDTA if doing IMAC)
- ~1 µl/ml Benzonase (to reduce viscosity) [54]
Cell Lysis:
- Harvest cells and wash with cold PBS.
- Resuspend cell pellet in 2-5 volumes of the freshly prepared lysis buffer.
- Incubate on a rotator for 15-30 minutes at 4°C to ensure complete lysis.
Clarification:
- Centrifuge the lysate at 18,000 x g for 30 minutes at 4°C to pellet insoluble material.
- Immediately transfer the clarified supernatant (the protein lysate) to a new pre-chilled tube.
- Proceed directly to downstream applications like immunoprecipitation or immunoblotting. Avoid repeated freeze-thaw cycles [54].
Quantitative Inhibitor Profiling
Selecting the right inhibitor requires understanding its potency and selectivity. The data below, synthesized from profiling studies, provides a comparative overview.
Table 3: Profiling Data for Common DUB Inhibitors
| Inhibitor | Primary Target(s) | Reported IC₅₀ / Potency | Key Selectivity Notes | Ideal Application in Lysis |
|---|---|---|---|---|
| PR-619 | Broad-spectrum, pan-DUB inhibitor (cysteine proteases) | ~10-20 µM (in vitro) [2] | Inhibits many USP, UCH, and OTU family DUBs. Not selective. | Global ubiquitinome stabilization for proteomics [14]. |
| VLX1570 | USP14 (preferentially), UCHL5 | IC₅₀ ~1.5-18 µM (USP14, SPR) [53] | Preferentially binds and stabilizes USP14. More selective than pan-inhibitors. | Studying proteasomal degradation or specific USP14 biology. |
| XL177A | USP7 | Nanomolar potency [29] | A highly selective, covalent inhibitor developed as a chemical probe. | Validating USP7-specific substrates and functions. |
Diagram 1: Inhibitor Selection Workflow
Diagram 2: Redox Regulation of DUB Activity
{ /article }
Within the broader context of preventing deubiquitination during cell lysis, managing the chemical environment is paramount for successful deubiquitinase (DUB) inhibitor research. A fundamental challenge arises from the interplay between essential reducing agents and the catalytic cysteine residues of DUBs. Many DUBs are cysteine proteases that rely on an active-site cysteine nucleophile for their enzymatic activity, making them prime targets for covalent inhibitor design [55] [56]. However, standard lysis buffers often contain reducing agents like dithiothreitol (DTT) or β-mercaptoethanol (BME) to maintain protein solubility and prevent artificial oxidation. These same agents can directly compromise the efficacy of cysteine-reactive DUB inhibitors, such as Ub-propargylamide (Ub-PA) and other activity-based probes (ABPs), by reducing the critical electrophilic warhead intended to form a covalent bond with the DUB [55] [56]. This technical guide addresses specific troubleshooting scenarios to help researchers navigate this complex biochemical balance.
Frequently Asked Questions (FAQs) & Troubleshooting
Q1: Why does my DUB inhibitor fail to work in my standard RIPA lysis buffer, even at high concentrations?
- Likely Cause: The most probable cause is the presence of a reducing agent in your lysis buffer. Reducing agents like DTT and β-mercaptoethanol are added to buffers to break disulfide bonds and keep proteins reduced and soluble [57]. However, they will also reduce the reactive electrophile (e.g., vinyl sulfone, propargylamide) on your DUB inhibitor, rendering it inert and unable to covalently modify the active-site cysteine of the target DUB [55].
- Solution:
- Prepare a fresh aliquot of lysis buffer without any reducing agent for experiments involving cysteine-reactive DUB inhibitors.
- If protein solubility is a concern without a reducing agent, consider switching to a non-ionic detergent-based lysis buffer like NP-40 or a modified RIPA without SDS and deoxycholate, which may be less harsh [58] [42].
- Always add the DUB inhibitor to the lysate after cell lysis is complete, as the intracellular environment also contains reducing molecules like glutathione.
Q2: I cannot omit reducing agents entirely from my lysis protocol as my target protein precipitates. What are my options?
- Likely Cause: Some transmembrane or multi-cysteine proteins require a reduced state to remain in solution [57].
- Solution:
- Titrate the reducing agent: Systematically lower the concentration of DTT or BME to find a minimal level that maintains target protein solubility but does not completely inactivate your inhibitor. Start with concentrations 10-fold lower than your standard protocol (e.g., 0.1 mM DTT instead of 1 mM).
- Use a more potent inhibitor: Some inhibitors have highly reactive warheads that can outcompete low levels of reducing agents. Consult the literature for inhibitors with faster kinetics.
- Switch inhibitors: If possible, investigate whether a non-covalent, allosteric, or non-cysteine-targeting DUB inhibitor is available for your DUB of interest.
- Change the order of operations: Ensure the inhibitor is given time to bind to the DUB before the lysate is exposed to the reducing agent. This can sometimes be achieved by pre-treating intact cells with a membrane-permeable inhibitor prior to lysis.
Q3: My activity-based probe (ABP) shows unexpected, non-catalytic labeling in my lysates. What could be happening?
- Likely Cause: Chemoproteomic studies have revealed that DUB ABPs can sometimes label non-catalytic cysteine residues on DUBs and even on non-DUB proteins, especially in complex lysates [55]. This is not necessarily an "activity-based" reaction and can lead to misinterpretation.
- Solution:
- Validate labeling sites: Employ reactive-site-centric chemoproteomic methods to confirm the probe is labeling the catalytic cysteine [55]. This involves using a cleavable biotin tag and LC-MS/MS to identify the exact modified residue.
- Use appropriate controls: Always include a catalytically inactive DUB mutant (e.g., Cys-to-Ala mutation) in your experiments. If the ABP labels the mutant with similar efficiency, the labeling is not activity-based [56].
- Optimize probe design: Bifunctional probes with an alkyne handle for downstream bioorthogonal tagging can help distinguish specific enrichment from non-specific binding [55].
Key Data and Experimental Protocols
Quantitative Impact of Reducing Agents on Common DUB Inhibitors
Table 1: Impact of Common Lysis Buffer Components on DUB Inhibitor Efficiency
| Lysis Buffer Component | Typical Concentration | Impact on Cysteine-Reactive DUB Inhibitors | Recommended Action |
|---|---|---|---|
| Dithiothreitol (DTT) | 1-10 mM | High Impact: Potently reduces electrophilic warheads, leading to complete inactivation [57]. | Omit or use at very low concentrations (<0.1 mM). |
| β-mercaptoethanol (BME) | 0.1-1% (v/v) | High Impact: Similar to DTT, can inactivate covalent inhibitors [57]. | Omit or use at low concentrations (<0.01%). |
| Triton X-100 / NP-40 | 0.1-1% (v/v) | Low Impact: Non-ionic detergents aid lysis and generally do not interfere with inhibitor chemistry [58] [42]. | Generally safe to use. |
| SDS | 0.1-1% (w/v) | Medium Impact: Ionic detergent can denature proteins, potentially altering DUB conformation and inhibitor binding site [58]. | Use milder detergents (e.g., NP-40) for native conditions. |
| Protease Inhibitors | As per cocktail | Low Impact: Essential for preventing protein degradation without affecting most inhibitor chemistries [59] [60]. | Always include, but confirm compatibility. |
Standard Protocol for Cell Lysis with DUB Inhibitors
This protocol is optimized for adherent mammalian cells to be used in conjunction with cysteine-reactive DUB inhibitors [59] [60] [42].
Materials:
- Ice-cold PBS (Phosphate Buffered Saline)
- Lysis Buffer (without reducing agents): 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% NP-40 (or Triton X-100), 1x Protease Inhibitor Cocktail (EDTA-free if concerned about metalloproteases), 1x Phosphatase Inhibitor Cocktail [60] [42].
- Your chosen DUB inhibitor, reconstituted in an appropriate solvent (e.g., DMSO).
- Cell scraper, microcentrifuge tubes, sonicator (optional).
Method:
- Grow and Wash Cells: Culture adherent cells to ~80% confluence. Aspirate the media and gently wash the cell monolayer twice with ice-cold PBS.
- Lyse Cells: Add ice-cold lysis buffer directly to the culture dish (e.g., 200-500 µL for a 60 mm dish). Swirl to distribute the buffer evenly.
- Scrape and Incubate: Use a chilled cell scraper to dislodge the cells and transfer the lysate to a pre-cooled microcentrifuge tube. Incubate on ice for 15-30 minutes with occasional vortexing.
- Clarify Lysate: Centrifuge the lysate at 13,000 - 16,000 x g for 15 minutes at 4°C to pellet insoluble debris.
- Add DUB Inhibitor: Critical Step: Transfer the clarified supernatant (soluble protein fraction) to a new tube. Now, add your DUB inhibitor from a concentrated stock and incubate for the desired time and temperature (e.g., 30 minutes at 37°C for active-site engagement).
- Proceed with Analysis: The lysate is now ready for downstream applications such as western blotting, immunoprecipitation, or activity assays.
Diagram 1: DUB Inhibitor Lysis Workflow
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagents for DUB Inhibitor Research
| Reagent / Material | Function / Role | Key Consideration |
|---|---|---|
| Cysteine-Reactive DUB Inhibitors (e.g., Ub-PA, Ub-VS) | Covalently modify the active-site cysteine of DUBs, allowing for detection, enrichment, and inhibition of active DUBs [55] [56]. | Highly susceptible to reduction; use with buffers free of DTT/BME. |
| Non-Ionic Detergent Lysis Buffers (e.g., NP-40, Triton X-100) | Disrupts lipid membranes to release soluble proteins while maintaining protein-protein interactions and native enzyme conformations [58] [42]. | Milder than RIPA; ideal for preserving DUB activity and inhibitor binding. |
| Protease & Phosphatase Inhibitor Cocktails (EDTA-free) | Prevents co-purifying proteases from degrading DUBs and phosphatases from altering their phosphorylation state during lysis [59] [60]. | EDTA-free versions are available if the DUB of interest is a metalloenzyme. |
| Activity-Based Probes (ABPs) | Chemical tools that report on the functional state of entire enzyme families in complex proteomes via a reactive group and a reporter tag [55]. | Confirm catalytic site labeling using chemoproteomic methods to avoid off-target artifacts [55]. |
| Bioorthogonal Tags (e.g., Alkyne handles) | Allow for click chemistry-based conjugation of reporter tags (like biotin or fluorophores) to ABPs after lysis and labeling, minimizing steric interference [55] [61]. | Enables highly specific detection and purification of probe-labeled proteins. |
Diagram 2: Cysteine Reactivity and Inhibition
Troubleshooting Guide
Q: My western blot for specific ubiquitin linkages (e.g., K48) shows a weak or absent signal after cell lysis, even with DUB inhibitors. What could be wrong? A: This is often due to incomplete inhibition of DUBs or improper lysis conditions.
- Solution 1: Verify the concentration and stability of your DUB inhibitor. Some inhibitors, like PR-619, are used at high concentrations (e.g., 10-50 µM). Ensure they are freshly prepared in DMSO and added to the lysis buffer immediately before use.
- Solution 2: Pre-chill all equipment and buffers. Perform lysis rapidly on ice to minimize DUB activity before inhibition is complete.
- Solution 3: Increase the stringency of your lysis buffer. Include 1% SDS and briefly boil samples to denature proteins and irreversibly inactivate DUBs, followed by dilution for immunoprecipitation.
Q: I see high background or non-specific bands in my linkage-specific ubiquitin blot. How can I improve specificity? A: Non-specific binding is common with ubiquitin antibodies.
- Solution 1: Optimize antibody dilution. A high concentration can lead to off-target binding. Perform a titration series (e.g., 1:500 to 1:5000) to find the optimal signal-to-noise ratio.
- Solution 2: Increase the stringency of your wash buffers after immunoprecipitation. Add 0.1% SDS or 500 mM NaCl to your standard RIPA or TBS-T wash buffers.
- Solution 3: Include a control using an isotype-matched IgG or a bead-only control to identify non-specific bands.
Q: My mass spectrometry data shows a low yield of ubiquitinated peptides, especially for specific linkages. How can I enhance detection? A: Sample preparation is critical for MS-based ubiquitinomics.
- Solution 1: Use a ubiquitin enrichment step. Utilize antibodies that pan-specifically recognize di-glycine (K-ε-GG) remnants left after trypsin digestion to enrich for ubiquitinated peptides.
- Solution 2: Employ Tandem Ubiquitin Binding Entities (TUBEs) during lysis. TUBEs protect ubiquitin chains from DUBs and can pull down a broad range of ubiquitinated proteins, increasing your starting material.
- Solution 3: Use a protease like Glu-C in addition to trypsin. Glu-C cleaves at different sites, generating longer peptides that can sometimes be easier to identify and quantify.
Frequently Asked Questions (FAQs)
Q: Why is it crucial to use a cocktail of DUB inhibitors rather than a single one? A: Different DUB inhibitor classes target specific DUB families. For example, PR-619 is a broad-spectrum inhibitor, while G5 targets USP-family DUBs, and MJD inhibitor targets Machado-Joseph Disease domain DUBs. Using a cocktail (e.g., 10 µM PR-619, 1 µM G5) ensures coverage across multiple DUB families, providing more comprehensive protection for all linkage types.
Q: How do I choose between different linkage-specific antibodies? A: The choice depends on application and validation.
- Western Blot: Look for antibodies validated for immunoblotting. Check the vendor's data for clear, single bands at the expected molecular weights.
- Immunofluorescence/Immunohistochemistry: Requires antibodies validated for these specific applications.
- Cross-reactivity: Always consult the vendor's datasheet for information on cross-reactivity with other Ub linkages or modified forms. Independent validation using ubiquitin mutants (e.g., K48-only, K63-only) is the gold standard.
Q: What are the key advantages of TUBEs over traditional DUB inhibitors? A: TUBEs offer a dual function: they act as high-affinity DUB inhibitors by sequestering ubiquitin chains from endogenous DUBs, and they simultaneously affinity-purify polyubiquitinated proteins. This makes them superior for downstream applications like mass spectrometry or studying endogenous ubiquitination without overexpression.
Data Presentation
Table 1: Efficacy of Common DUB Inhibitors in Preserving Ubiquitin Linkages
| Inhibitor Name | Target DUB Family | Typical Working Concentration | Efficacy for K48 | Efficacy for K63 | Efficacy for K11 | Key Consideration |
|---|---|---|---|---|---|---|
| PR-619 | Broad-spectrum | 10 - 50 µM | High | High | High | Can be toxic to cells; use for lysis only. |
| G5 | USP-family | 1 - 5 µM | Moderate | Moderate | Moderate | Often used in a cocktail with other inhibitors. |
| PYR-41 (E1 Inhibitor) | Ubiquitin Activating Enzyme | 10 - 50 µM | High | High | High | Blocks all ubiquitination; use for "deubiquitination" controls. |
| TUBEs (Agarose beads) | N/A (Binds chains) | 10 - 50 µg | Very High | Very High | Very High | Also enriches ubiquitinated proteins; ideal for proteomics. |
Table 2: Comparison of Ubiquitin Linkage-Specific Antibodies
| Linkage Type | Vendor A (Cat#) | Vendor B (Cat#) | Recommended Application | Reported Cross-reactivity |
|---|---|---|---|---|
| K48 | Abcam (ab140601) | Cell Signaling (8081) | WB, IP | Minimal with K63, K11 |
| K63 | Millipore (05-1308) | CST (5621) | WB, IF, IHC | Minimal with K48, K11 |
| K11 | Millipore (ABS77) | Abcam (ab134957) | WB, IP | Can vary; validate carefully. |
Experimental Protocols
Protocol 1: Cell Lysis for Ubiquitin Chain Preservation
Objective: To extract proteins while preserving endogenous ubiquitin chains of all linkage types. Reagents:
- Lysis Buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 0.5% Sodium Deoxycholate, 0.1% SDS, 1 mM EDTA.
- DUB Inhibitor Cocktail: 25 µM PR-619, 1 µM G5, 10 mM N-Ethylmaleimide (NEM).
- Protease Inhibitor Cocktail (without EDTA).
- Phosphatase Inhibitor Cocktail.
- PBS (ice-cold).
Procedure:
- Pre-chill microcentrifuge tubes and lysis buffer on ice.
- Prepare fresh lysis buffer and supplement with DUB inhibitor cocktail, protease, and phosphatase inhibitors immediately before use.
- Aspirate media from cultured cells and wash once with ice-cold PBS.
- Lyse cells directly on the culture plate by adding an appropriate volume of lysis buffer (e.g., 100 µL per 1x10^6 cells). Scrape cells and transfer the lysate to a pre-chilled microcentrifuge tube.
- Vortex briefly and incubate on ice for 15-30 minutes with occasional vortexing.
- Centrifuge at 14,000 x g for 15 minutes at 4°C to pellet insoluble material.
- Carefully transfer the supernatant (whole-cell lysate) to a new pre-chilled tube. Proceed immediately to protein quantification and downstream analysis (e.g., Western blot, IP).
Protocol 2: Immunoprecipitation of Ubiquitinated Proteins for Western Blot Analysis
Objective: To isolate ubiquitinated proteins for specific detection of linkage types. Reagents:
- Protein A/G Agarose Beads.
- Linkage-specific ubiquitin antibody (e.g., anti-K48-Ub) and control IgG.
- Lysis Buffer (as in Protocol 1).
- Wash Buffer: Lysis buffer without SDS.
Procedure:
- Pre-clear 500 µg of whole-cell lysate by incubating with 20 µL of Protein A/G beads for 1 hour at 4°C with end-over-end rotation. Centrifuge briefly and transfer supernatant to a new tube.
- Add 1-2 µg of linkage-specific ubiquitin antibody or control IgG to the pre-cleared lysate. Incubate for 2 hours at 4°C with rotation.
- Add 30 µL of Protein A/G beads and incubate for an additional 1-2 hours (or overnight) at 4°C with rotation.
- Pellet beads by gentle centrifugation (2000 x g for 2 min) and carefully aspirate the supernatant.
- Wash the beads 3-4 times with 1 mL of ice-cold Wash Buffer, resuspending completely each time.
- After the final wash, completely remove the supernatant.
- Elute proteins by adding 2X Laemmli sample buffer and boiling at 95°C for 10 minutes. Analyze by Western blot.
Mandatory Visualizations
Diagram Title: Ubiquitin Preservation Workflow
Diagram Title: Ubiquitin Linkage Functional Roles
The Scientist's Toolkit
Table 3: Essential Reagents for Ubiquitin Chain Validation
| Reagent | Function | Example |
|---|---|---|
| Broad-Spectrum DUB Inhibitor | Irreversibly inhibits a wide range of DUBs during lysis to prevent chain disassembly. | PR-619 |
| USP-Family DUB Inhibitor | Specifically targets Ubiquitin-Specific Proteases, often used in inhibitor cocktails. | G5 |
| Alkylating Agent | Modifies cysteine residues, inactivating cysteine-dependent DUBs. | N-Ethylmaleimide (NEM) |
| Tandem Ubiquitin Binding Entities (TUBEs) | High-affinity ubiquitin-binding molecules that protect chains from DUBs and aid in enrichment. | Agarose-TUBE (LifeSensors) |
| Linkage-Specific Ub Antibodies | Detect and immunoprecipitate polyubiquitin chains formed via specific lysine residues. | Anti-K48-Ub (CST #8081) |
| Di-Glycine (K-ε-GG) Antibody | Enriches for ubiquitinated peptides for mass spectrometry analysis by recognizing the tryptic remnant. | Anti-K-ε-GG (CST #5562) |
| Proteasome Inhibitor | Prevents degradation of ubiquitinated proteins by the proteasome, increasing their steady-state level. | MG-132 |
Welcome to the Technical Support Center for Deubiquitinase (DUB) Research. This resource addresses the critical challenge of preventing premature deubiquitination during cell lysis, a fundamental step for accurate analysis of ubiquitin signaling. The content below provides targeted troubleshooting guides and FAQs to help researchers account for cell-type-specific DUB expression profiles in their experimental designs.
Frequently Asked Questions (FAQs)
FAQ 1: Why is it crucial to consider cell type when designing cell lysis protocols for ubiquitination studies? Different cell types express varying levels and types of Deubiquitinating Enzymes (DUBs). During cell lysis, the loss of cellular compartmentalization brings substrates and DUBs into contact, creating an artificial environment where deubiquitination can rapidly occur. If not inhibited, these active DUBs can strip ubiquitin chains from your protein of interest, leading to inaccurate representation of its true ubiquitination state in vivo. The high expression of specific DUBs in certain cancers, for instance, necessitates a more aggressive inhibition strategy [62] [63] [64].
FAQ 2: What is the most common mechanism of DUB inhibition during lysis? The most common mechanism involves targeting the catalytic cysteine residue present in the active site of most DUB families (USP, UCH, OTU, MJD). DUBs are cysteine proteases, and their activity relies on a catalytic triad that lowers the pKa of this cysteine, making it prone to oxidation and modification [5]. Alkylating agents like N-Ethylmaleimide (NEM) or Iodoacetamide are widely used to covalently modify this catalytic cysteine, permanently inactivating the enzyme.
FAQ 3: I am using a high concentration of DUB inhibitors, but still observe loss of ubiquitin signal. What could be wrong? This is a common problem with several potential causes:
- Incomplete Inhibition: The chosen inhibitor concentration may be insufficient for the specific DUB profile of your cell line. Some DUBs are more resistant to certain inhibitors.
- Oxidative Inactivation of Inhibitors: Reactive Oxygen Species (ROS) in the lysate can reversibly oxidize the catalytic cysteine of DUBs, forming sulphenyl-amide intermediates. While this inactivates the DUB, the modification is reversible upon reduction. If your lysis buffer contains reducing agents like DTT or β-mercaptoethanol, they can reactivate these DUBs, leading to post-lysis deubiquitination [5]. Always add DUB inhibitors before and ensure they are present during lysis.
FAQ 4: How does the structural rigidity of my source material impact lysis efficiency? The chosen lysis method must be appropriate for your cell type's physical barriers.
- Mammalian Cells: These have only a plasma membrane and are relatively easy to lyse with detergent-based buffers.
- Bacterial Cells: These have a rigid peptidoglycan cell wall. Gram-negative bacteria have an additional outer membrane, making them more resistant to lysis and often requiring mechanical disruption or enzymatic pre-treatment with lysozyme [17] [65].
- Plant Cells: These possess a tough cellulose cell wall, making physical methods like grinding with a mortar and pestle under liquid nitrogen the most effective approach [66].
Troubleshooting Guides
Problem: Inconsistent Ubiquitination Results Across Different Cell Lines
Potential Cause: Variations in the expression levels and subtypes of DUBs between cell lines.
Solutions:
- Profile DUB Expression: Before experiments, use public databases (e.g., The Cancer Genome Atlas) or perform Western blotting to identify which DUBs are highly expressed in your cell models. For example, USP5 is often upregulated in non-small cell lung cancer (NSCLC) and pancreatic cancer [67].
- Use a Pan-DUB Inhibitor Cocktail: Instead of a single inhibitor, use a broad-spectrum agent like PR-619. PR-619 is a cell-permeable, broad-range DUB inhibitor that has been shown to induce the accumulation of ubiquitinated proteins and subsequent ER stress in oesophageal squamous cell carcinoma cells [2].
- Optimize Lysis Buffer Composition: Supplement standard RIPA buffer with a cocktail of specific and pan-DUB inhibitors. See Table 1 for recommended reagents.
Problem: High Background or Protein Degradation in Lysates
Potential Cause: Inefficient lysis leading to prolonged processing or release of proteases/DUBs from subcellular compartments.
Solutions:
- Pre-chill Equipment and Work Quickly: Perform all steps at 4°C to slow enzymatic activity.
- Choose the Correct Lysis Method: Match the physical lysis method to your cell type. Sonication is highly efficient for bacterial and yeast cells, while a Dounce homogenizer may be better for cultured mammalian cells [65] [66].
- Include Protease Inhibitors: Always use a broad-spectrum protease inhibitor cocktail in conjunction with DUB inhibitors.
- Add Nuclease: To reduce lysate viscosity from released DNA/RNA, add DNase/RNase, or use sonication which shears genomic DNA [66].
Essential Research Reagent Solutions
Table 1: Key Reagents for Preventing Deubiquitination During Cell Lysis
| Reagent | Function/Mechanism | Example & Usage |
|---|---|---|
| Pan-DUB Inhibitors | Broad-spectrum inhibition of multiple DUB families by targeting the catalytic cysteine. | PR-619: Used in the 10-50 µM range in cell culture media or lysis buffers. Shown to induce ubiquitin-protein aggregation and ER stress [2]. |
| Specific DUB Inhibitors | Target individual DUBs or specific subfamilies; useful for validating roles of specific DUBs. | EOAI: A USP5 inhibitor. Studies in NSCLC used it to induce DNA damage and apoptosis, highlighting its utility in cancer models with USP5 upregulation [67]. |
| Alkylating Agents | Irreversibly modify cysteine residues, inactivating DUBs and other cysteine-dependent enzymes. | N-Ethylmaleimide (NEM): Commonly used at 5-25 mM in lysis buffers. Must be prepared fresh. |
| Metal Chelators | Inhibit JAMM/MPN+ family metalloprotease DUBs (e.g., BRCC36, AMSH) by chelating zinc. | 1,10-Phenanthroline: Used at 1-10 mM concentration. |
| Protease Inhibitors | Inhibit serine, cysteine, aspartic, and metallo-proteases to prevent general protein degradation. | Commercial Cocktails (e.g., PMSF, Leupeptin, Aprotinin): Used per manufacturer's instructions. |
Protocol: Preventing Deubiquitination During Lysis of Adherent Mammalian Cells
Materials:
- Pre-chilled PBS
- Lysis Buffer (e.g., RIPA): Supplemented with fresh 10-20 µM PR-619 and 10 mM NEM.
- Cell Scraper
- Microcentrifuge
Method:
- Prepare Lysis Buffer: Add PR-619 and NEM to ice-cold lysis buffer immediately before use. Do not add reducing agents.
- Wash Cells: Aspirate culture medium and wash cells once with pre-chilled PBS.
- Lyse Cells: Add an appropriate volume of supplemented lysis buffer directly to the culture dish (e.g., 100-200 µL for a 35 mm dish).
- Harvest Lysate: Using a cell scraper, quickly scrape the cells off the dish and transfer the lysate to a pre-chilled microcentrifuge tube.
- Incubate: Maintain constant agitation for 15-30 minutes at 4°C.
- Clarify: Centrifuge at >12,000 x g for 15 minutes at 4°C to pellet insoluble debris.
- Collect Supernatant: Transfer the clarified supernatant (total cell lysate) to a new pre-chilled tube. Protein concentration can now be determined, and reducing agents can be added for downstream applications like SDS-PAGE.
Quantitative Data on DUB Inhibitor Effects
Table 2: Documented Effects of DUB Inhibition in Various Cancer Cell Lines
| Cell Line | Cancer Type | DUB Target / Inhibitor | Key Phenotypic Outcome | Reference |
|---|---|---|---|---|
| Oesophageal Squamous Cell Carcinoma (ESCC) | Oesophageal Cancer | Pan-DUB / PR-619 | Induced G2/M cell cycle arrest, apoptosis, and autophagy via ER stress. | [2] |
| A549, H460 | Non-Small Cell Lung Cancer (NSCLC) | USP5 / EOAI | Induced DNA damage, p53 activation, cell cycle arrest, and apoptosis. Synergized with cisplatin. | [67] |
| Various (e.g., NSCLC, HCC) | Multiple Cancers | Multiple DUBs (e.g., JOSD2, CSN5, USP29) | Promoted aerobic glycolysis (Warburg effect) by stabilizing metabolic enzymes (ALDOA, PFK1, HK2) and transcription factors (MYC, HIF1α). | [64] |
Signaling Pathways & Workflow Visualizations
DUB Redox Regulation Mechanism
This diagram illustrates how Reactive Oxygen Species (ROS) can reversibly inactivate DUBs, a key consideration for buffer composition.
Experimental Workflow for DUB-Inhibited Lysis
This flowchart outlines the critical steps for a successful cell lysis procedure that preserves ubiquitination.
In research focused on deubiquitination and deubiquitinase (DUB) inhibitors, maintaining the authentic ubiquitin landscape during cell lysis is paramount. The rapid and promiscuous activity of DUBs once cells are disrupted can artificially alter protein ubiquitination states, leading to experimental artifacts that compromise data interpretation. This guide addresses common troubleshooting challenges within the context of preventing deubiquitination during cell lysis, providing actionable solutions for researchers, scientists, and drug development professionals.
FAQ: Addressing Deubiquitination-Specific Artifacts
1. What causes protein smearing in western blots when studying DUB inhibition?
Protein smearing in western blots, particularly when analyzing ubiquitinated proteins, is frequently caused by unspecific protein aggregation and incomplete inhibition of deubiquitination during sample preparation.
- Mechanism: Broad-spectrum DUB inhibitors containing α,β-unsaturated carbonyl groups (e.g., b-AP15, VLX1570) can act as non-specific Michael acceptors. Rather than selectively inhibiting DUBs, they can cross-link a wide range of cellular proteins, leading to high molecular weight aggregates that appear as smears on blots [68].
- Solution: Switch to more selective DUB inhibitors or probe mechanisms beyond DUB inhibition. Validate findings using genetic DUB knockdown approaches to confirm that observed effects are truly due to DUB inhibition rather than compound-mediated cross-linking artifacts. Ensure lysis buffers contain adequate concentrations of proven DUB inhibitors and are supplemented with fresh protease inhibitors.
2. Why is there a loss of signal for specific ubiquitin chains in my assays?
Loss of specific ubiquitin chain signal often results from incomplete DUB inhibition during cell lysis, allowing residual DUB activity to cleave ubiquitin chains before analysis.
- Mechanism: Different DUB subfamilies (USP, UCH, OTU, MJD) have varying sensitivities to chemical inhibitors. If your inhibitor cocktail does not adequately target all relevant DUB classes present in your sample, specific ubiquitin linkages may be preferentially degraded during the lysis procedure [29].
- Solution: Optimize DUB inhibitor cocktails by including compounds targeting multiple DUB subfamilies. As an example, PR-619 (a broad-spectrum DUB inhibitor) has been shown to induce accumulation of ubiquitylated proteins and subsequent endoplasmic reticulum stress, demonstrating effective pan-DUB inhibition [2]. Always prepare fresh lysis buffers with DUB inhibitors immediately before use and ensure rapid processing of samples post-lysis.
3. What leads to inconsistent results between replicates in DUB inhibitor studies?
Inconsistency typically stems from variability in cell lysis efficiency and inconsistent DUB inhibitor activity across samples.
- Mechanism: Mechanical lysis methods (e.g., sonication, homogenization) can generate heat, potentially denaturing proteins and inactivating DUB inhibitors if not properly controlled. Additionally, the timing between lysis and complete denaturation of proteins can vary, allowing different degrees of deubiquitination across replicates [66].
- Solution: Standardize lysis protocols with precise timing. Use pre-chilled equipment and maintain samples on ice throughout processing. Consider chemical lysis methods with optimized buffers for more reproducible results across replicates [69]. Validate lysis efficiency and consistency by measuring protein yield and integrity across replicates.
Troubleshooting Guide: Experimental Artifacts and Solutions
Table 1: Common Artifacts and Their Solutions in Deubiquitination Research
| Artifact Type | Potential Causes | Recommended Solutions |
|---|---|---|
| Protein Smearing | Non-specific protein cross-linking by Michael acceptor inhibitors [68]; Protein aggregation due to ubiquitinated protein accumulation [2] | Use more selective DUB inhibitors; Include aggregation-reducing agents in lysis buffer; Lower sample loading on gels |
| Loss of Ubiquitin Signal | Incomplete DUB inhibition during lysis; Degradation of ubiquitin chains by active DUBs; Improper lysis buffer composition | Optimize DUB inhibitor cocktail concentration; Validate inhibitor efficacy; Flash-freeze samples before lysis; Use denaturing lysis buffers when appropriate |
| Inconsistent Results | Variable lysis efficiency between samples; Inconsistent heating during sonication; Degraded DUB inhibitors | Standardize lysis timing and protocol; Use chemical lysis methods for reproducibility [69]; Prepare fresh inhibitor stocks aliquots |
Optimized Experimental Protocols
Protocol 1: Preventing Deubiquitination During Cell Lysis for Western Blotting
This protocol is designed to preserve the native ubiquitinome by effectively inhibiting DUB activity during cell lysis.
Reagents and Materials:
- Pre-chilled DUB-Inhibited Lysis Buffer (see Reagent Table below)
- Broad-spectrum DUB inhibitor (e.g., PR-619 at 25-50 µM) [2]
- Protease Inhibitor Cocktail (without EDTA)
- PMSF (1 mM) or other serine protease inhibitor
- Benzonase (25-50 µg/mL) to reduce viscosity [66]
- Pre-chilled PBS
- Mechanical homogenizer (Dounce or Potter-Elvehjem) or sonicator
Procedure:
- Preparation: Pre-chill centrifuge, homogenizer, and microcentrifuge tubes on ice. Prepare fresh DUB-Inhibited Lysis Buffer immediately before use.
- Cell Harvesting: Wash cells with ice-cold PBS and quickly aspirate completely.
- Lysis: Add appropriate volume of lysis buffer (typically 100-200 µL per 10⁶ cells) directly to cell pellet.
- Immediate Mixing: Vortex vigorously for 10-15 seconds to ensure rapid inhibitor penetration.
- Mechanical Disruption: Perform 10-15 strokes with pre-chilled Dounce homogenizer or pulse sonicate (3 × 10-second pulses at 30% amplitude) on ice.
- Incubation: Incubate on ice for 10 minutes with occasional vortexing.
- Clarification: Centrifuge at 14,000 × g for 15 minutes at 4°C.
- Collection: Immediately transfer supernatant to new pre-chilled tube and proceed to protein quantification or add Laemmli buffer for western blotting.
Critical Steps:
- The time between buffer addition and complete lysis should be minimized to <5 minutes.
- Always include a control without DUB inhibitors to confirm their necessity.
- Avoid repeated freeze-thaw cycles of lysates; instead, aliquot and store at -80°C.
Protocol 2: DUB Inhibitor Validation Using Activity-Based Protein Profiling (ABPP)
This protocol uses ABPP to validate DUB inhibitor efficacy and selectivity in cellular extracts, ensuring your inhibitors are functioning as intended [29].
Reagents and Materials:
- Biotinylated ubiquitin-based probes (e.g., Ub-VME or Ub-PA)
- Streptavidin-conjugated beads
- DUB inhibitors for testing
- HEK293T or other relevant cell line
- Lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1% NP-40)
Procedure:
- Prepare Cellular Extracts: Lyse cells in appropriate buffer without DUB inhibitors.
- Inhibitor Pretreatment: Incubate cell extracts with DUB inhibitors (e.g., 50 µM) or DMSO control for 30 minutes at room temperature.
- ABP Labeling: Add biotinylated ubiquitin probes to extracts and incubate for an additional 60 minutes.
- Streptavidin Pulldown: Capture probe-labeled DUBs using streptavidin beads.
- Analysis: Elute proteins and analyze by western blotting for specific DUBs or by mass spectrometry for proteome-wide profiling.
Validation Metrics:
- Successful DUB inhibition is indicated by reduced ABP labeling of target DUBs.
- Selectivity is confirmed by minimal off-target effects on other DUBs or unrelated enzymes.
The Scientist's Toolkit: Essential Reagents for DUB Research
Table 2: Key Research Reagents for Deubiquitination Studies
| Reagent Category | Specific Examples | Function and Application |
|---|---|---|
| Broad-Spectrum DUB Inhibitors | PR-619 [2] | Pan-DUB inhibitor; induces ubiquitinated protein accumulation and ER stress; useful for initial target validation |
| Selective DUB Inhibitors | XL177A (USP7-selective) [29] | Targets specific DUB family members; enables precise pharmacological interrogation of individual DUB functions |
| Activity-Based Probes | Biotin-Ub-VME, Biotin-Ub-PA [29] | Covalently label active DUBs; essential for profiling DUB activity and inhibitor selectivity in native systems |
| Chemical Library Frameworks | N-cyanopyrrolidines, Azetidine-based compounds [29] | Provide starting points for development of selective DUB inhibitors through rational library design |
| Lysis Additives | Benzonase, Lysozyme, Protease Inhibitor Cocktails [66] | Reduce viscosity and prevent protein degradation during extraction; crucial for maintaining sample integrity |
Experimental Workflows and Signaling Pathways
DUB Inhibitor Mechanism and Validation Workflow
DUB Inhibition Signaling Pathway
Successfully troubleshooting artifacts in deubiquitination research requires a comprehensive understanding of DUB biology coupled with meticulous attention to experimental details during cell lysis. By implementing the standardized protocols, validation methods, and reagent strategies outlined in this guide, researchers can significantly enhance the reliability and reproducibility of their findings. The integration of ABPP for inhibitor validation and careful selection of DUB inhibitors based on their mechanisms of action provides a robust framework for generating high-quality data in the increasingly important field of DUB research and drug development.
Assessing Inhibition Efficacy and Benchmarking Method Performance
The integrity of ubiquitination states is paramount in proteomics research and drug development. A primary challenge during cell lysis is the rapid, unwanted activity of deubiquitinating enzymes (DUBs), which can strip proteins of their ubiquitin tags, thereby erasing critical regulatory information. This guide provides targeted troubleshooting and foundational protocols for key analytical methods—Western Blotting, Activity-Based Probes (ABPs), and Mass Spectrometry—to help researchers successfully preserve ubiquitin signatures by integrating DUB inhibitors into their workflow.
Western Blotting Troubleshooting for Ubiquitinated Proteins
Western blotting is essential for detecting ubiquitinated proteins, but researchers often face specific challenges. The table below outlines common issues and their solutions.
| Problem Scenario | Possible Cause | Recommended Solution |
|---|---|---|
| No or weak signal | Poor transfer efficiency of high molecular weight ubiquitin chains [70] [71] | Pre-equilibrate gel in transfer buffer with 0.02-0.04% SDS; use 0.2 µm pore size PVDF membrane [70]. |
| Low protein concentration or ubiquitination level [71] | Increase protein load; use high-quality DUB inhibitors in lysis buffer to preserve ubiquitin marks. | |
| High background | Inadequate blocking or non-specific antibody binding [71] | Optimize blocking with 5% BSA or non-fat dry milk; increase number/duration of washes with Tween-20 [71]. |
| Non-specific bands | Antibody cross-reactivity [71] | Use validated, specific antibodies; pre-adsorb antibodies with blocking agent; verify DUB inhibitor specificity. |
| Smearing or diffuse bands | Protein degradation during lysis [70] | Ensure fresh DUB inhibitors and complete protease inhibitor cocktails are in lysis buffer; keep samples cold. |
| Uneven or swirling bands | Poor gel-to-membrane contact or air bubbles [70] | Roll a glass pipette over membrane during assembly to remove bubbles; ensure proper saturation of filter pads [70]. |
Key Experimental Protocol: Western Blotting for Ubiquitin
- Cell Lysis: Lyse cells in RIPA buffer supplemented with 5-10 µM of broad-spectrum DUB inhibitors (e.g., PR-619) and 1x protease inhibitor cocktail. Keep samples on ice and clarify by centrifugation [72] [35].
- Gel Electrophoresis: Load 20-50 µg of protein onto a 4-12% Bis-Tris gel. High-percentage gels are better for resolving polyubiquitin chains [70].
- Transfer: For proteins >100 kDa, pre-equilibrate gel for 10 minutes in transfer buffer with 0.02% SDS. Transfer to a 0.2 µm PVDF membrane using a wet or semi-dry system [70].
- Blocking and Antibody Incubation: Block membrane with 5% BSA in TBST for 1 hour. Incubate with primary anti-ubiquitin antibody (1:1000) overnight at 4°C, followed by HRP-conjugated secondary antibody for 1 hour [71].
- Detection: Develop using enhanced chemiluminescence (ECL) substrate and image.
Activity-Based Probes for DUB Analysis
Activity-based probes (ABPs) are powerful tools for directly monitoring the functional state of DUBs in complex biological samples. They covalently bind to the active site of DUBs, reporting on their activity rather than mere abundance [35] [73].
Troubleshooting DUB ABPs
| Problem Scenario | Possible Cause | Recommended Solution |
|---|---|---|
| No labeling in lysates | DUBs not active or probe degraded [73] | Confirm DUB activity; use fresh ABP; include positive control lysate. |
| No labeling in live cells | Probe is cell impermeable [35] | Use a cell-permeable small-molecule ABP (e.g., with BODIPY or Cy dye) [35] [73]. |
| High background labeling | Probe concentration too high or non-specific binding [73] | Titrate ABP to optimal concentration; include a competitive inhibitor control. |
| Incomplete DUB inhibition | DUB inhibitor is ineffective or unstable [72] | Use fresh, potent inhibitors; validate inhibition with Ub-AMC assay. |
Key Experimental Protocol: Profiling DUB Activity with ABPs
- Probe Design: A typical DUB ABP has three components:
- Reactive Group (Warhead): An electrophile (e.g., vinyl methyl ester -VME) that covalently binds the catalytic cysteine of DUBs [35] [73].
- Recognition Element: Ubiquitin or a small-molecule scaffold that directs the probe to DUB active sites [35].
- Tag: A reporter (e.g., biotin for purification, or a fluorophore like Cy5 for detection) [73].
- In Vitro Labeling (Lysates): Incubate 50 µg of cell lysate (prepared with minimal detergent) with 1 µM ABP for 1 hour at 37°C. Stop reaction with SDS-PAGE loading buffer [73].
- In Situ Labeling (Live Cells): Incubate intact cells with a cell-permeable ABP (e.g., 5 µM) for 2-4 hours. Wash cells, harvest, and lyse for analysis [35].
- Detection: Analyze by in-gel fluorescence if using a fluorescent probe, or by streptavidin blot if using a biotinylated probe.
Mass Spectrometry for Ubiquitinomics
Mass spectrometry (MS) is the gold standard for system-wide mapping of ubiquitination sites. Preventing deubiquitination during sample preparation is critical for accurate data [74] [75].
Troubleshooting MS in Ubiquitinomics
| Problem Scenario | Possible Cause | Recommended Solution |
|---|---|---|
| Few ubiquitin peptides identified | Sample complexity too high [74] | Fractionate samples using high-pH reversed-phase chromatography to reduce complexity [74]. |
| Deubiquitination during preparation [72] | Include DUB inhibitors in all lysis and digestion buffers; work quickly on ice. | |
| Poor reproducibility | Inconsistent sample preparation [74] | Use standardized MS sample prep kits (e.g., EasyPep); quantify peptides before LC-MS [74]. |
| High background in blanks | Contaminated buffers or system [76] | Prepare fresh buffers; use clean, dedicated plastics; flush LC system [76]. |
Key Experimental Protocol: Sample Preparation for Ubiquitinomics MS
- Cell Lysis and Digestion: Lyse cells in a denaturing buffer (e.g., 8 M Urea) containing 5-10 µM DUB inhibitors. Reduce disulfide bonds with DTT, alkylate with iodoacetamide, and digest proteins with trypsin [74] [75].
- Peptide Cleanup and Fractionation: Desalt peptides using C18 solid-phase extraction. To increase depth of coverage, fractionate peptides using a high-pH reversed-phase spin column [74].
- LC-MS/MS Analysis: Analyze peptides on a Q-TOF or Orbitrap mass spectrometer coupled to a nanoflow LC system. Use data-dependent acquisition (DDA) to fragment and identify peptides.
- Data Analysis: Search MS/MS data against a protein database using software like MaxQuant or Proteome Discoverer, specifying ubiquitin remnant motif (Gly-Gly on lysine) as a variable modification [74].
The Scientist's Toolkit: Research Reagent Solutions
| Reagent / Material | Function in Experiment | Key Consideration |
|---|---|---|
| Broad-spectrum DUB Inhibitors (e.g., PR-619) | Inhibits a wide range of cysteine-dependent DUBs during cell lysis [72] [35]. | Use at 5-10 µM in lysis buffer; can be toxic to live cells. |
| Activity-Based Probes (e.g., HA-Ub-VME) | Covalently labels active DUBs for functional profiling in lysates [35] [73]. | Cell impermeable; use in lysates for accurate DUBome activity snapshot. |
| Cell-Permeable ABPs (e.g., BODIPY-labeled) | Profiles DUB activity in live, intact cells, preserving cellular context [35] [73]. | Allows for target engagement studies and in-cell visualization. |
| PVDF Membrane (0.2 µm) | Membrane for Western blotting; retains proteins, especially small or large ubiquitin chains [70]. | Superior to nitrocellulose for protein retention; pre-wet with methanol [70]. |
| High-pH Reversed-Phase Fractionation Kit | Reduces sample complexity for deep ubiquitinome analysis by MS [74]. | Increases number of quantifiable peptides and protein identifications. |
| Quantitative Fluorometric Peptide Assay | Accurately quantifies peptides before LC-MS analysis to ensure equal loading [74]. | Improves reproducibility and quantitative accuracy in MS experiments. |
Frequently Asked Questions (FAQs)
Q1: Why is it critical to include DUB inhibitors in my cell lysis buffer for ubiquitination studies? Deubiquitinating enzymes remain active during cell lysis and can rapidly remove ubiquitin chains from protein substrates within minutes. This erases the ubiquitination signal you are trying to measure. Including broad-spectrum DUB inhibitors in your lysis buffer immediately upon cell disruption is essential to "freeze" the native ubiquitin state of the proteome [72] [35].
Q2: What is the main advantage of using ABPs over traditional Western blotting for studying DUBs? Activity-Based Probes report on the functional state of DUBs—specifically, the fraction that is active at a given time. Western blotting with DUB antibodies only measures protein abundance. Since DUB activity is highly regulated by conformational changes, protein interactions, and post-translational modifications, ABPs provide a more physiologically relevant picture of the "DUBome" [35] [73].
Q3: I get a high background in my Western blots for ubiquitin. What are the first steps to troubleshoot this? First, optimize your blocking conditions by using 5% BSA and increasing the blocking time. Second, increase the number and duration of washes with TBST containing 0.1% Tween-20. Third, titrate your primary and secondary antibodies, as excessive antibody concentration is a common cause of high background [71].
Q4: My mass spectrometry experiment identified very few ubiquitinated peptides. Where should I look to improve my results? The two most critical areas to check are sample preparation and fractionation. First, ensure DUB inhibitors were present from the moment of lysis to prevent degradation. Second, fractionate your peptide sample using a high-pH reversed-phase kit before LC-MS/MS. This reduces sample complexity, allowing the instrument to sequence more low-abundance ubiquitinated peptides [74].
Q5: How can I study DUB activity in live cells rather than in lysates? Traditional Ub-based ABPs are too large to cross the cell membrane. To profile DUBs in live cells, you need to use recently developed cell-permeable, small-molecule ABPs. These probes contain a smaller recognition element and a fluorescent tag (like BODIPY or Cy dye) that allows them to enter cells and label active DUBs in their native cellular environment [35] [73].
Ubiquitin chain cleavage assays are fundamental tools for quantifying the activity of deubiquitinating enzymes (DUBs) in complex biological lysates. These assays enable researchers to investigate the specificity, kinetics, and regulation of DUBs, which are crucial regulators of protein stability, signaling pathways, and cellular homeostasis. Within the context of thesis research focused on preventing deubiquitination during cell lysis, robust quantitative assessment of DUB activity becomes paramount for validating inhibition strategies and ensuring accurate measurement of cellular ubiquitination states. This technical support center provides comprehensive troubleshooting guides and detailed methodologies to address the specific challenges researchers encounter when implementing these assays, particularly when working with lysates where preserving native ubiquitin conjugates is essential.
The following diagram illustrates the core conceptual and experimental workflow for conducting DUB activity assays in lysates, highlighting the critical step of DUB inhibition during cell lysis within the broader experimental context.
Essential Concepts and Preparation
Fundamental Principles of Ubiquitin Chain Biology
Ubiquitination is a post-translational modification where ubiquitin, a 76-amino acid protein, is attached to substrate proteins. This process involves a sequential enzyme cascade: E1 (activating), E2 (conjugating), and E3 (ligase) enzymes [77]. Ubiquitin itself contains seven lysine residues (K6, K11, K27, K29, K33, K48, and K63) and an N-terminal methionine that can form polyubiquitin chains with distinct biological functions [78] [77]. The specific linkage type and chain length create a sophisticated "ubiquitin code" that determines the fate of modified proteins, most notably targeting them for proteasomal degradation (typically K48-linked chains) or regulating non-proteolytic processes like DNA repair and signal transduction (often K63-linked chains) [77].
Deubiquitinating enzymes (DUBs) are proteases that reverse ubiquitination by cleaving ubiquitin from modified substrates. They regulate ubiquitin-dependent signaling pathways, maintain free ubiquitin pools, and process ubiquitin precursors [79]. The human genome encodes approximately 100 DUBs categorized into several families based on their catalytic domains, with the ubiquitin-specific proteases (USPs) being the largest family [80]. DUBs exhibit varying degrees of specificity toward different ubiquitin chain linkage types and lengths, which determines their biological functions [81].
Preparation of DUB Inhibitors for Cell Lysis
Preventing deubiquitination during cell lysis is critical for preserving the native ubiquitin landscape in experimental systems. The following table summarizes common DUB inhibitors used in lysis buffers:
Table 1: Common DUB Inhibitors for Cell Lysis Preparation
| Inhibitor | Final Concentration | Specificity | Mechanism of Action | Stability in Lysis Buffer |
|---|---|---|---|---|
| PR-619 | 10-50 µM | Broad-spectrum | Inhibits multiple DUB families by reversible covalent modification | Stable for 24 hours at 4°C |
| N-Ethylmaleimide (NEM) | 5-10 mM | Cysteine proteases | Alkylates catalytic cysteine residues in multiple DUB families | Stable but must be added fresh before use |
| Ubiquitin Aldehyde | 1-5 µM | Ubiquitin-binding site | Mimics the ubiquitin C-terminus and binds active sites of many DUBs | Moderate stability; freeze aliquots |
| TLCK | 100-200 µM | Some DUB families | Serine protease inhibitor that also affects certain DUB classes | Stable for several weeks at -20°C |
Preparation Guidelines:
- Prepare concentrated stock solutions in appropriate solvents (DMSO for PR-619, water for NEM)
- Add inhibitors to chilled lysis buffer immediately before use
- Maintain samples on ice throughout lysis procedure
- Avoid repeated freeze-thaw cycles of inhibitor stocks
- Include control lysates without inhibitors to assess efficacy
Quantitative Assay Optimization
Substrate Selection and Preparation
Defined ubiquitin chains of specific linkages and lengths are essential substrates for quantitative DUB assays. Recombinant tetra-ubiquitin chains (K48-linked or K63-linked) have become the gold standard for measuring DUB activity in mutational analyses [78]. The length of ubiquitin chains significantly impacts their recognition by ubiquitin-binding proteins, including DUBs, making chain length a critical parameter in assay design [81].
Preparation of Defined Ubiquitin Chain Substrates:
- Source recombinant ubiquitin chains from commercial suppliers or express and purify in-house
- Verify linkage specificity using linkage-specific antibodies or mass spectrometry [82]
- Confirm chain length by gel electrophoresis and mass spectrometry
- Quantify accurately using spectrophotometry (A280 measurement)
- Prepare aliquots to avoid repeated freeze-thaw cycles
For specialized applications, branched ubiquitin chains can be utilized, as certain DUBs like UCH37 show marked preference for these architectures [83]. The development of ubiquitin-absolute quantification/parallel reaction monitoring (Ub-AQUA/PRM) mass spectrometry methods now allows direct and highly sensitive measurement of the stoichiometry of all eight ubiquitin-ubiquitin linkage types simultaneously [82] [84].
Experimental Protocol: DUB Activity Assay in Lysates
Materials Required:
- Cell lysates prepared with DUB inhibitors
- Defined ubiquitin chain substrates (e.g., K48-linked tetra-ubiquitin)
- Reaction buffer (50 mM Tris-HCl pH 7.5, 50 mM NaCl, 5 mM DTT, 0.1 mg/mL BSA)
- Stop solution (4X SDS-PAGE loading buffer with 8% β-mercaptoethanol)
- Pre-cast gels (10-20% gradient) for ubiquitin separation
- Immunoblotting equipment and linkage-specific ubiquitin antibodies
Step-by-Step Procedure:
- Prepare reaction mixtures on ice containing:
- 1-10 µg of cell lysate protein
- 200-500 ng of ubiquitin chain substrate
- Reaction buffer to final volume of 20-50 µL
Include essential controls:
- No-lysate control (substrate only)
- No-substrate control (lysate only)
- Boiled-lysate control (heat-inactivated DUBs)
- Specific DUB inhibitor controls
Initiate reactions by transferring to 37°C and incubate for appropriate timepoints (e.g., 0, 5, 15, 30, 60 minutes)
Stop reactions by adding stop solution and heating at 95°C for 5 minutes
Separrate cleavage products by SDS-PAGE using 10-20% gradient gels
Transfer proteins to PVDF membrane and immunoblot with linkage-specific ubiquitin antibodies
Quantify band intensities using densitometry software, measuring both substrate depletion and product formation
Quantitative Data Analysis
Key Parameters to Calculate:
- Reaction velocity: Initial rate of substrate cleavage (pmol/min)
- Specific activity: Reaction velocity normalized to total lysate protein (pmol/min/µg)
- Kinetic constants: Km and kcat if using multiple substrate concentrations
- Inhibition efficacy: Percentage reduction in activity compared to untreated controls
Table 2: Troubleshooting Quantitative Measurements in DUB Assays
| Problem | Potential Causes | Solutions | Quantitative Impact |
|---|---|---|---|
| High background cleavage | Incomplete DUB inhibition during lysis | Optimize inhibitor cocktail; shorten lysis time; maintain low temperature | Can overestimate basal DUB activity by 30-70% |
| Non-linear reaction kinetics | Substrate depletion; enzyme instability; product inhibition | Use initial rate measurements; reduce lysate amount; shorter time courses | Invalidates kinetic calculations if not addressed |
| Variable lysate activity | Differences in cell number; lysis efficiency; protein quantification | Normalize to cell count; use standardized lysis protocols; verify protein assays | Can cause 2-3 fold variations between replicates |
| Poor antibody sensitivity | Inappropriate antibody dilution; low-affinity antibodies; transfer issues | Optimize antibody concentrations; validate antibodies with controls; check transfer efficiency | Reduces dynamic range and quantitative accuracy |
| Inconsistent substrate quality | Substrate degradation; improper storage; freeze-thaw cycles | Fresh aliquot for each experiment; quality control assessment; proper storage at -80°C | Can completely invalidate comparative experiments |
Troubleshooting Guides and FAQs
Common Experimental Challenges and Solutions
FAQ 1: How can I prevent unintended deubiquitination during cell lysis?
- Implement a comprehensive DUB inhibitor cocktail in your lysis buffer
- Perform lysis at 4°C or on ice with pre-chilled buffers
- Minimize lysis time and process samples quickly
- Validate inhibition efficacy by comparing with and without inhibitors
- Consider using alternative lysis methods (e.g., mechanical disruption) that proceed more rapidly
FAQ 2: What are the best controls for DUB activity assays in lysates? Essential controls include:
- No-substrate control (detects endogenous ubiquitin signals)
- No-lysate control (assesses non-enzymatic substrate breakdown)
- Heat-inactivated lysate (controls for non-specific cleavage)
- Specific inhibitor controls (validates enzyme-specific activity)
- Time-zero timepoint (established baseline cleavage)
FAQ 3: Why do I see multiple cleavage products in my assay?
- Many DUBs process ubiquitin chains sequentially rather than endonucleolytically
- Lysates contain multiple DUBs with different cleavage specificities
- Some DUBs generate intermediate fragments with distinct migration patterns
- Consider using chain-terminating ubiquitin mutants (e.g., K48R) to distinguish specific cleavage activities
FAQ 4: How can I distinguish between different DUB activities in complex lysates?
- Use linkage-specific substrates (e.g., K48- vs K63-linked chains)
- Implement specific DUB inhibitors or activators when available
- Combine with immunodepletion of specific DUBs before assay
- Employ catalytic mutants as negative controls in overexpression systems
FAQ 5: What quantification method provides the most accurate DUB activity measurements? Mass spectrometry-based approaches like Ub-AQUA/PRM provide the most precise quantification of ubiquitin chain topology and cleavage [82] [84]. However, for routine assessment, densitometric analysis of immunoblots with careful standard curves offers a practical balance between accuracy and accessibility. Fluorescence-based assays using labeled ubiquitin provide real-time kinetics but may alter enzyme-substrate interactions.
Advanced Technique: Mass Spectrometry-Based Quantification
For thesis-level research requiring high-precision quantification, targeted mass spectrometry methods offer superior accuracy for assessing ubiquitin chain topology and DUB cleavage patterns [84]. The Ub-AQUA/PRM (ubiquitin-absolute quantification/parallel reaction monitoring) methodology enables direct and highly sensitive measurement of the stoichiometry of all eight ubiquitin-ubiquitin linkage types simultaneously [82].
Key Steps in MS-Based DUB Assay Quantification:
- Stabilize ubiquitin chains biochemically immediately after cleavage reactions
- Digest with specific proteases (e.g., trypsin) to generate signature peptides
- Spike with heavy isotope-labeled internal standard peptides
- Analyze by LC-PRM/MS with optimized transitions for each linkage type
- Quantify using standard curves generated from synthetic heavy peptides
This approach is particularly valuable for characterizing DUBs with unique specificities, such as those targeting branched ubiquitin chains [83] or exhibiting chain length preferences [81].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for DUB Activity Assays
| Reagent Category | Specific Examples | Research Application | Technical Considerations |
|---|---|---|---|
| Defined Ubiquitin Chains | K48-linked tetra-ubiquitin, K63-linked di-ubiquitin | Substrate specificity profiling; kinetic analysis | Verify linkage purity; assess chain length homogeneity; proper storage at -80°C |
| DUB Inhibitors | PR-619, N-Ethylmaleimide (NEM), Ubiquitin aldehyde | Preservation of ubiquitin conjugates during lysis; control experiments | Prepare fresh stocks; optimize concentration for specific lysates; consider cytotoxicity |
| Linkage-Specific Antibodies | Anti-K48 ubiquitin, Anti-K63 ubiquitin, Anti-linear ubiquitin | Detection of specific cleavage products; immunoblot analysis | Validate specificity with defined chains; optimize dilution for different applications |
| Activity-Based Probes | HA-Ub-VS, HA-Ub-Br2, Cy5-labeled ubiquitin derivatives | DUB profiling; active site labeling; competition studies | Use controlled labeling conditions; include appropriate controls for specificity |
| Recombinant DUBs | USP5, USP39, UCH37, OTUB1 | Positive controls; substrate specificity mapping | Confirm catalytic activity with fluorogenic substrates before use |
| Mass Spectrometry Standards | Heavy isotope-labeled ubiquitin signature peptides | Absolute quantification of ubiquitin chain topology | Use stable isotope-labeled internal standards for precise quantification |
The following diagram illustrates the strategic application of these research reagents throughout the experimental workflow for assessing DUB activity in lysates.
The ubiquitin-proteasome system (UPS) represents one of the most important regulatory mechanisms for intracellular protein homeostasis, controlling protein degradation, localization, and function [14]. Within this system, deubiquitinating enzymes (DUBs) perform the essential function of removing ubiquitin from substrate proteins, thereby reversing the action of E3 ubiquitin ligases [32]. During cell lysis and protein extraction, the normal regulation of DUB activity is disrupted, leading to potential experimental artifacts through unwanted deubiquitination of protein substrates. This deubiquitination can alter protein stability, function, and degradation signals, ultimately compromising experimental results.
Preventing unwanted deubiquitination during cell lysis is therefore paramount for researchers studying ubiquitin-dependent processes, including protein turnover, DNA damage response, signal transduction, and immune regulation [14] [32]. The selection of appropriate DUB inhibitor cocktails becomes a critical methodological consideration that can significantly impact data quality and interpretation. This technical support document provides a comprehensive analysis of commercial DUB inhibitor options, their efficacy profiles, and practical implementation strategies to support researchers in maintaining ubiquitin homeostasis during experimental procedures.
Research Reagent Solutions: Essential Materials for DUB Inhibition
Table 1: Key Research Reagents for DUB Inhibition Studies
| Reagent Category | Specific Examples | Function & Application |
|---|---|---|
| Broad-Spectrum DUB Inhibitors | PR619 [14] | Pan-DUB inhibitor; useful for proteomic studies to globally stabilize ubiquitinated substrates |
| Proteasome-Associated DUB Inhibitors | Auranofin [85] | Specifically targets proteasome-associated DUBs UCHL5 and USP14; gold-containing compound |
| Activity-Based Probes | HA-Ub-PA, HA-Ub-VME, Biotin-UbVMe [86] [4] | Covalently label active DUBs for detection, enrichment, or inhibition assessment |
| Commercial Protease Inhibitor Cocktails | TargetMol Protease Inhibitor Cocktail [87], Roche Protease Inhibitor Cocktail [88] | Broad-spectrum protease inhibition with varying efficacy against DUBs |
| Selective DUB Inhibitors | XL177A (USP7 inhibitor) [32], FT671 (USP7 inhibitor) [86] | Highly specific inhibitors for individual DUB target validation |
Comparative Analysis of Commercial DUB Inhibitor Cocktails
Composition and Efficacy Profiles
Table 2: Quantitative Comparison of Commercial DUB Inhibitor Cocktails
| Product Feature | TargetMol Protease Inhibitor Cocktail [87] | Roche Protease Inhibitor Cocktail [88] |
|---|---|---|
| Primary Composition | AEBSF, Aprotinin, Bestatin, E-64, Leupeptin, Pepstatin A | Proprietary formulation (exact components not specified) |
| Serine Protease Inhibition | AEBSF (104 mM), Aprotinin (80 µM) | Effective |
| Cysteine Protease Inhibition | E-64 (1.5 mM), Leupeptin (2 mM) | Effective against some cysteine proteases |
| Aspartic Protease Inhibition | Pepstatin A (1.5 mM) | Not specifically mentioned |
| Aminopeptidase Inhibition | Bestatin (5 mM) | Not specifically mentioned |
| Efficacy Against DUBs | Varies by DUB type; ineffective against some DUBs like Ataxin-3 [87] | Ineffective against MDV UL36-DUB [88] |
| Inhibition Mechanism | Combination of irreversible (AEBSF, E-64) and reversible inhibitors | Not specified |
| Compatible Applications | Western Blot, Co-IP, Pull-down, IF, IHC, Kinase assays [87] | Not specified in available literature |
Experimental Evidence for Differential Efficacy
Research has demonstrated significant variability in the effectiveness of commercial inhibitor cocktails against different DUB families. A key study on Marek's disease virus-encoded UL36 deubiquitinase (UL36-DUB) revealed striking differences in inhibitor efficacy, finding that "UL36-DUB exhibits resistance to the Roche protease inhibitor cocktail and serine protease inhibitor, but not to the Solarbio protease inhibitor cocktail" [88]. This finding highlights the critical importance of matching specific DUB targets with appropriate inhibition strategies.
Furthermore, manufacturers acknowledge limitations in DUB coverage, with TargetMol explicitly noting that "some DUB proteases (one example is ATAXIN-3) cannot be suppressed by traditional protease inhibitors, such as E-64, AEBSF, bestatin, leupeptin and Aprotinin" [87]. This transparency helps researchers make informed decisions based on their specific DUB targets.
Troubleshooting Guides & FAQs
Frequently Asked Questions
Q1: Why does my commercial protease inhibitor cocktail fail to prevent deubiquitination during cell lysis? A: Most standard protease inhibitor cocktails are optimized for common proteases like trypsin, chymotrypsin, and papain, but may lack efficacy against specific DUB families due to structural variations in DUB active sites [88] [87]. The resistance of certain DUBs to conventional inhibitors necessitates targeted approaches.
Q2: How can I validate the effectiveness of my DUB inhibition strategy? A: Utilize activity-based probes (ABPs) such as HA-Ub-VME or Biotin-UbVMe [4] to directly monitor DUB activity in lysates. These covalent probes label active DUBs and can be detected via Western blot or mass spectrometry to confirm inhibition efficacy.
Q3: Are there specific DUB families particularly resistant to standard inhibitors? A: Yes, research indicates significant variation in DUB susceptibility. For example, the viral UL36-DUB shows resistance to some commercial cocktails [88], and human Ataxin-3 (a Machado-Joseph disease domain DUB) is known to resist common cysteine protease inhibitors [87].
Q4: What negative controls should I include when testing DUB inhibitors? A: Always include samples without DUB inhibitors to establish baseline deubiquitination, and consider using catalytically inactive DUB mutants or siRNA knockdowns where possible to confirm specificity of observed effects.
Troubleshooting Common Experimental Issues
Problem: Incomplete deubiquitination prevention despite using commercial cocktails
- Potential Cause: The cocktail may not target your specific DUB due to active site variations.
- Solution: Supplement with broad-spectrum DUB inhibitors like PR-619 [14] or investigate target-specific inhibitors such as Auranofin for proteasome-associated DUBs [85].
Problem: High background in ubiquitin detection assays
- Potential Cause: Inconsistent inhibitor concentration or degradation of inhibitor components.
- Solution: Prepare fresh inhibitor cocktails for each experiment, verify concentration optimization through dose-response experiments, and include activity probes to monitor DUB inhibition in real-time [4].
Problem: Cellular toxicity from inhibitor cocktails
- Potential Cause: Some DUB inhibitors, particularly metal-containing compounds like Auranofin, can affect multiple cellular pathways [85].
- Solution: Titrate inhibitor concentration to find the minimum effective dose, reduce exposure time where possible, and validate findings with multiple inhibitor classes.
Problem: Inconsistent results between experimental replicates
- Potential Cause: Variability in cell lysis conditions or incomplete inhibition during preparation.
- Solution: Standardize lysis protocols, pre-chill all equipment and buffers, and add inhibitors immediately upon cell disruption.
Methodologies and Experimental Protocols
Standard Protocol for Cell Lysis with DUB Inhibition
DUB Inhibition During Cell Lysis Workflow
Preparation of Inhibitor Cocktail:
Cell Lysis Procedure:
- Harvest cells and wash with ice-cold phosphate-buffered saline (PBS) [86].
- Resuspend cell pellet in lysis buffer (50 mM Tris, 5 mM MgCl₂, 0.5 mM EDTA, 250 mM sucrose, 1 mM DTT, pH 7.5) containing fresh protease and DUB inhibitors [86].
- Lyse using preferred method (mechanical disruption, detergent-based, etc.) while maintaining samples at 4°C throughout the process.
- Clarify lysates by centrifugation at 14,000 × g for 25 minutes at 4°C [86].
- Determine protein concentration and proceed immediately to downstream applications.
Validation Protocol for DUB Inhibition Efficacy
Activity-Based Probe Labeling:
Ubiquitinome Stabilization Assessment:
Technical Specifications and Application Guidelines
DUB Inhibitor Mechanisms and Spectral Coverage
DUB Inhibitor Coverage and Limitations
The effectiveness of DUB inhibition strategies varies significantly based on the chemical properties of the inhibitors and the structural characteristics of target DUBs. Commercial cocktails like the TargetMol formulation provide broad coverage against standard protease classes through multiple mechanisms [87]:
- AEBSF (104 mM): Irreversible serine protease inhibitor targeting trypsin-like proteases
- E-64 (1.5 mM): Irreversible cysteine protease inhibitor effective against papain-family enzymes
- Leupeptin (2 mM): Reversible inhibitor of both cysteine and serine proteases
- Pepstatin A (1.5 mM): Reversible aspartic protease inhibitor targeting pepsin and cathepsin D
- Bestatin (5 mM): Reversible aminopeptidase inhibitor affecting multiple metalloproteases
Despite this comprehensive coverage, certain DUBs remain resistant to these conventional inhibitors due to unique active site architectures or mechanistic features [88] [87]. This limitation necessitates customized approaches for specific DUB families or research applications requiring complete ubiquitinome stabilization.
Based on current evidence, researchers should approach DUB inhibition with the understanding that standard protease inhibitor cocktails provide incomplete protection against deubiquitination during cell lysis. The selection of appropriate inhibitors should be guided by several factors:
Target Specificity: When studying specific DUBs, investigate their sensitivity to different inhibitor classes through preliminary experiments or literature review.
Application Requirements: For global ubiquitinome studies, supplement standard cocktails with broad-spectrum DUB inhibitors like PR-619 [14]. For specific pathways, consider targeted inhibitors like Auranofin for proteasomal DUBs [85].
Validation Imperative: Always include activity-based profiling to confirm DUB inhibition efficacy in your specific experimental system [4].
Cost-Benefit Analysis: While specialized DUB inhibitors may increase reagent costs, this investment is justified by significantly improved data quality and reliability in ubiquitination studies.
The field continues to evolve with new inhibitor technologies emerging regularly, particularly activity-based probes and selective small-molecule inhibitors that promise more targeted and effective DUB suppression strategies [15] [89].
PR-619 is a small-molecule, broad-spectrum inhibitor of deubiquitinating enzymes (DUBs), which are cysteine proteases that cleave ubiquitin from protein substrates [90]. Its primary application in research is to inhibit DUB activity during experiments, thereby preventing the deubiquitination of target proteins and leading to the accumulation of ubiquitinated proteins within cells [91] [90]. This property makes it a valuable chemical tool for investigating the roles of ubiquitination in various cellular processes, including protein degradation, signal transduction, and autophagy [14].
Molecular Characteristics: PR-619, also known as 2,6-Diaminopyridine-3,5-bis(thiocyanate), has a molecular weight of 223.3 g/mol and is typically reconstituted in DMSO for use in cell-based studies [90].
Experimental Protocol: Monitoring Ubiquitinated Protein Accumulation
This protocol details the method for validating PR-619 efficacy by monitoring the accumulation of polyubiquitinated proteins in cell cultures.
Reagent Preparation
- PR-619 Stock Solution (10 mM): Reconstitute 1 mg of lyophilized PR-619 powder in 0.45 mL of pure DMSO [90].
- Storage: Store the lyophilized powder at -20°C (desiccated) where it is stable for 24 months. Once reconstituted, the stock solution should be aliquoted, stored at -20°C, and used within one month to prevent loss of potency. Avoid multiple freeze-thaw cycles [90].
- Working Concentration: The final working concentration in cell culture media can vary but is often in the range of 5-20 µM to observe DUB inhibition. Note: Concentrations above 20 µM have been reported to induce DNA topoisomerase II (TOP2) poisoning, an off-target effect that should be considered in experimental design [91].
Cell Treatment and Lysis
- Cell Seeding: Plate cells in an appropriate culture vessel and allow them to adhere and reach the desired confluence (e.g., 60-80%).
- Treatment: Treat cells with the predetermined working concentration of PR-619 (e.g., 5-20 µM) for a specified duration. Common treatment times range from 3 to 6 hours, though kinetics experiments show effects can be observed within 10 minutes [14]. A control group should be treated with an equivalent volume of DMSO vehicle.
- Cell Lysis: After treatment, lyse the cells using a suitable lysis buffer (e.g., RIPA buffer) supplemented with protease inhibitors and, critically, DUB inhibitors other than PR-619 (e.g., N-ethylmaleimide) to preserve the ubiquitination state during and after lysis.
- Centrifugation: Clarify the cell lysates by centrifugation at high speed (e.g., 14,000 × g for 15 minutes at 4°C). Collect the supernatant for subsequent analysis.
Detection and Analysis
- Immunoblotting (Western Blot): This is the standard method for detecting the accumulation of polyubiquitinated proteins.
- Separate equal amounts of protein lysate by SDS-PAGE.
- Transfer the proteins to a nitrocellulose or PVDF membrane.
- Probe the membrane with an anti-polyubiquitin antibody (e.g., FK1, FK2, or P4D1) or an antibody that detects the di-Glycine remnant (K-ε-GG) after tryptic digestion, which is specific for ubiquitinated proteins [14].
- An increase in the high-molecular-weight smear on the immunoblot in PR-619-treated samples compared to the DMSO control indicates successful accumulation of polyubiquitinated proteins.
- Quantification: Densitometric analysis of the ubiquitin smear or specific ubiquitinated bands can be performed to quantify the extent of accumulation.
The workflow can be summarized as follows:
Data Presentation: Quantitative Effects of PR-619
The following tables summarize key quantitative data from published research on PR-619, providing benchmarks for expected experimental outcomes.
Table 1: Ubiquitinated Protein Accumulation Kinetics with PR-619 Treatment
| Cell Line / System | PR-619 Concentration | Treatment Duration | Observed Effect | Citation |
|---|---|---|---|---|
| U2OS (His10-Ub) | Not Specified | 10, 30, 60, 180 min | Robust accumulation of ubiquitin conjugates observed at all time points [14]. | [14] |
| U2OS (His10-Ub) | Not Specified | 1, 3 hours (with TAK243) | Reduction in ubiquitin conjugates, indicating rapid DUB-mediated turnover [14]. | [14] |
| HEK293 Cell Extracts | 50 µM | Not Specified (ABPP screen) | Competitively blocked ABP labeling for 45+ endogenous DUBs, confirming broad target engagement [26]. | [26] |
Table 2: Functional Consequences of PR-619 Treatment in Various Models
| Experimental Model | PR-619 Concentration | Key Functional Outcome | Citation |
|---|---|---|---|
| Oligodendroglial Cell Line | Not Specified | Buildup of protein aggregates and increased expression of heat shock proteins [90]. | [90] |
| Urothelial Carcinoma Cells | Not Specified | Relieved chemoresistance to cisplatin [90]. | [90] |
| Retinal Ganglion Cells (RGCs) | 8.23 µM (in vivo) | Increased RGC survival in a glaucomatous model; upregulated parkin expression and enhanced mitophagy [38]. | [38] |
| General Cell-Based Studies | 5-20 µM | Concentration range for robust cellular DUB inhibitor activity [91]. | [91] |
The Scientist's Toolkit: Key Research Reagents
Table 3: Essential Reagents for PR-619 Validation Experiments
| Reagent / Material | Function / Description | Example / Note |
|---|---|---|
| PR-619 | Broad-spectrum DUB inhibitor; leads to accumulation of polyubiquitinated proteins. | Reconstitute in DMSO to 10 mM stock [90]. |
| DMSO (Vehicle Control) | Solvent for PR-619; used in control treatments. | Use the same volume as in PR-619-treated samples. |
| Protease Inhibitor Cocktail | Prevents general protein degradation during cell lysis. | Add to lysis buffer. |
| Additional DUB Inhibitors (e.g., NEM) | Prevents deubiquitination during cell lysis, preserving ubiquitin signals. | Critical for lysis buffer composition. |
| Anti-Polyubiquitin Antibody | Primary antibody for detecting polyubiquitinated proteins via Western blot. | e.g., FK1, FK2, or P4D1. |
| Anti-K-ε-GG Antibody | Antibody for enrichment or detection of ubiquitinated peptides in mass spectrometry. | Specific for tryptic diGly remnant [14]. |
| Proteasome Inhibitor (e.g., MG132) | Positive control; inhibits proteasomal degradation, also causing ubiquitin accumulation. | Can be used in combination studies [14]. |
| E1 Inhibitor (e.g., TAK243) | Negative control; inhibits ubiquitin activation, depleting ubiquitin conjugates [14]. |
Troubleshooting Guide & FAQs
Frequently Asked Questions
Q1: I am not observing an increase in the high-molecular-weight ubiquitin smear in my Western blot after PR-619 treatment. What could be wrong?
- A: This could be due to several factors:
- Insufficient inhibitor concentration or duration: Titrate the concentration of PR-619 (try 5, 10, 20 µM) and extend the treatment time.
- Instability of reconstituted PR-619: Ensure the stock solution is fresh, has been stored correctly at -20°C in aliquots, and has not undergone multiple freeze-thaw cycles [90].
- Deubiquitination during lysis: Confirm that your lysis buffer contains additional DUB inhibitors (e.g., N-ethylmaleimide) to prevent artifactural deubiquitination after cell disruption.
- Antibody sensitivity: Verify that your anti-ubiquitin antibody is suitable for Western blotting and try a different antibody if necessary.
- A: This could be due to several factors:
Q2: Are the effects of PR-619 reversible?
- A: PR-619 is a covalent DUB inhibitor. Its effects are generally not reversible upon washout, as it forms a stable, covalent bond with the catalytic cysteine of DUBs. The turnover of the inhibited DUBs and synthesis of new enzyme is required to restore activity.
Q3: What are the critical off-target effects I should be aware of when using PR-619?
- A: A key off-target effect identified is that PR-619 acts as a potent DNA topoisomerase II (TOP2) poison at concentrations similar to or above those used for DUB inhibition (above 20 µM) [91]. This can induce TOP2-DNA covalent complexes and confound results in studies related to DNA damage and repair. It is crucial to use the lowest effective concentration and include appropriate controls to distinguish DUB-related phenotypes from TOP2-related effects [91].
Q4: How does PR-619 compare to other DUB inhibitors like MG132?
- A: PR-619 and MG132 target different components of the ubiquitin system. PR-619 directly inhibits DUBs, preventing the removal of ubiquitin. MG132 is a proteasome inhibitor that blocks the degradation of ubiquitinated proteins. Both lead to an accumulation of polyubiquitinated proteins, but through distinct mechanisms. They can be used in combination for a synergistic accumulation of ubiquitin conjugates [14].
The relationship between PR-619's primary and off-target mechanisms is illustrated below:
For researchers investigating the ubiquitin-proteasome system (UPS), preserving the native ubiquitination state of proteins during cell lysis is a fundamental technical challenge. The dynamic and reversible nature of ubiquitination, mediated by deubiquitinating enzymes (DUBs), means that protein ubiquitination states can be dramatically altered within moments of cell disruption if proper precautions are not implemented. This technical support center provides comprehensive guidelines, troubleshooting advice, and standardized protocols to help researchers establish robust quality control metrics for successful ubiquitin preservation, framed within the broader context of DUB inhibitor research. Implementing these practices ensures that experimental results accurately reflect cellular ubiquitination states rather than artifacts introduced during sample preparation.
Fundamentals of Ubiquitination and DUB Activity
The Ubiquitin-Proteasome System
Protein ubiquitination is a post-translational modification involving a sequential enzymatic cascade that attaches ubiquitin molecules to target proteins. E1 activating enzymes, E2 conjugating enzymes, and E3 ligases work in concert to attach ubiquitin to substrate proteins, while DUBs selectively remove these modifications [6] [92]. This reversible process regulates diverse cellular functions including protein degradation, cell signaling, DNA repair, and immune responses [93] [94].
Deubiquitinating Enzymes: Challenges for Preservation
DUBs are specialized proteases that cleave ubiquitin from modified substrates or disassemble ubiquitin chains. The human genome encodes approximately 100 DUBs, which are classified into cysteine proteases (USPs, UCHs, OTUs, MJDs, MINDY, and ZUFSP) and metalloproteases (JAMM) [6] [94]. These enzymes maintain ubiquitin homeostasis, process ubiquitin precursors, and edit ubiquitin signals by removing ubiquitin molecules from target proteins [6] [95]. During cell lysis, the loss of compartmentalization and regulation allows DUBs to rapidly remove ubiquitin modifications unless they are properly inhibited.
Table: Major DUB Families and Their Characteristics
| DUB Family | Enzyme Type | Representative Members | Key Characteristics |
|---|---|---|---|
| USP | Cysteine protease | USP7, USP14 | Largest subfamily; diverse substrate recognition |
| UCH | Cysteine protease | UCH-L1, UCH-L3 | Prefer small ubiquitin adducts |
| OTU | Cysteine protease | OTUB1, A20 | Linkage-specific preferences |
| MJD | Cysteine protease | Ataxin-3, JOSD1 | Machado-Joseph disease domain |
| MINDY | Cysteine protease | MINDY-1, MINDY-2 | Prefer K48-linked ubiquitin chains |
| ZUFSP | Cysteine protease | ZUFSP/ZUP1 | Specific for K63-linked chains |
| JAMM | Metalloprotease | Rpn11, AMSH | Zinc-dependent; distinct mechanism |
Comprehensive Experimental Protocols
Standardized Lysis Buffer Formulation for Ubiquitin Preservation
Principle: The foundation of successful ubiquitin preservation is a properly formulated lysis buffer that simultaneously inactivates DUBs while preventing proteasomal degradation of ubiquitinated proteins.
Reagents Required:
- Base buffer (e.g., RIPA, NP-40, or Tris-based)
- DUB inhibitors (NEM, PR-619, or specific DUB inhibitors)
- Proteasome inhibitors (MG132, bortezomib, or carfilzomib)
- EDTA/EGTA
- Protease inhibitor cocktail (without DUB inhibitors)
- Phosphatase inhibitors (if studying phospho-ubiquitin)
Protocol Steps:
- Prepare Base Lysis Buffer: Start with 1 mL of your standard lysis buffer appropriate for your experimental system.
- Add EDTA/EGTA: Supplement with 5-10 mM EDTA or EGTA to chelate metal ions required by JAMM metalloprotease DUBs [96].
- Add DUB Inhibitors: Include 50-100 mM N-ethylmaleimide (NEM) to inhibit cysteine-based DUBs. For comprehensive inhibition, add 10-50 μM PR-619, a broad-spectrum DUB inhibitor [2] [93] [96].
- Add Proteasome Inhibitors: Include 10-25 μM MG132 or equivalent proteasome inhibitor to prevent degradation of ubiquitinated proteins during lysis [96] [92].
- Add Additional Inhibitors: Supplement with standard protease inhibitor cocktail (excluding DUB inhibitors) and phosphatase inhibitors if needed.
- Chill Buffer: Pre-cool complete lysis buffer to 4°C before use.
- Rapid Processing: Lyse cells directly in pre-cooled buffer and process immediately without delay.
Critical Note: NEM concentrations below 10 mM are insufficient for preserving K63-linked ubiquitin chains, which are particularly sensitive to DUB activity. Higher concentrations (50-100 mM) are required for comprehensive preservation of all ubiquitin chain types [96].
Quality Control Assessment Methods
Western Blot Analysis for Ubiquitin Preservation:
- Gel Selection: Use 8% Tris-glycine gels for optimal separation of long ubiquitin chains (>8 ubiquitin units) or 12% gels for better resolution of shorter chains (2-5 ubiquitin units) [96].
- Buffer System: Employ MOPS buffer for analyzing long ubiquitin chains (>8 units) or MES buffer for smaller chains (2-5 units) [96].
- Transfer Conditions: Use PVDF membranes with 0.2 μm pore size and transfer at 30V for 2.5 hours to prevent unfolding of ubiquitin chains [96].
- Antibody Selection: Carefully select ubiquitin antibodies based on recognition specificity. Many commercial antibodies show variable recognition of different linkage types [96].
Ubiquitin Enrichment and Pull-down Assays:
- Ubiquitin-Trap Methodology: Use ChromoTek Ubiquitin-Trap agarose or magnetic beads for immunoprecipitation of ubiquitinated proteins [92].
- Binding Capacity Consideration: Account for variable binding due to different ubiquitin chain lengths in quantitative experiments.
- Inhibitor Preservation: Maintain DUB inhibitors throughout all wash steps to prevent deubiquitination during enrichment.
Troubleshooting Guides & FAQs
Common Experimental Issues and Solutions
Table: Troubleshooting Common Ubiquitin Preservation Problems
| Problem | Potential Causes | Solutions |
|---|---|---|
| Weak or absent ubiquitin signal | Inadequate DUB inhibition; Improper inhibitor concentrations | Increase NEM to 50-100 mM; Verify MG132 activity; Use fresh PR-619 |
| High background smearing on western blots | Non-specific antibody binding; Incomplete transfer | Optimize antibody concentrations; Extend transfer time to 2.5 hours at 30V |
| Loss of specific ubiquitin linkages | Linkage-specific DUB activity; Antibody specificity issues | Use higher NEM concentrations for K63 chains; Validate antibody linkage specificity |
| Inconsistent results between replicates | Variable lysis times; Inhibitor degradation | Standardize lysis protocol across replicates; Prepare fresh inhibitors daily |
| Poor ubiquitin trap efficiency | Insufficient inhibitor during IP; Bead saturation | Add DUB inhibitors to IP wash buffers; Reduce input protein to avoid saturation |
Frequently Asked Questions
Q1: Why are both DUB inhibitors and proteasome inhibitors required in lysis buffers?
DUB inhibitors prevent the removal of ubiquitin signals by deubiquitinating enzymes, while proteasome inhibitors block the degradation of ubiquitinated proteins by the proteasome. These are complementary mechanisms - without proteasome inhibition, K48-linked polyubiquitinated proteins may be degraded during lysis, while without DUB inhibition, ubiquitin chains may be disassembled [96] [92].
Q2: What is the optimal duration for MG132 pre-treatment before lysis?
Research indicates that relatively short treatments (1-2 hours) with 5-25 μM MG132 are typically sufficient to preserve ubiquitination. Extended treatments (12-24 hours) can induce cellular stress responses and alter ubiquitination patterns, potentially confounding results [96].
Q3: How can I verify that my ubiquitin preservation methods are working effectively?
Include positive controls such as:
- Treatment with known DUB inhibitors (PR-619) to observe accumulated ubiquitin signals
- Comparison with E1 inhibitor (TAK243) treatment to deplete ubiquitin conjugates
- Use of linkage-specific ubiquitin antibodies to verify preservation of specific chain types
- Assessment of known ubiquitinated proteins in your system [93]
Q4: Can I use DUB inhibitors for in vivo experiments or only in cell culture?
While this guide focuses on cell lysis, many DUB inhibitors can be used in live cells to study DUB functions. However, considerations of cell permeability, toxicity, and specificity must be addressed for in vivo applications [2] [95].
Q5: Why does ubiquitin often appear as a smear rather than discrete bands on western blots?
Ubiquitinated proteins form a heterogeneous population with varying numbers of ubiquitin molecules (adding ~8 kDa per ubiquitin) attached to different lysine residues on target proteins. This molecular weight heterogeneity appears as a characteristic smear on western blots, which is actually indicative of successful preservation of diverse ubiquitination states [96] [92].
The Scientist's Toolkit: Essential Research Reagents
Table: Key Reagents for Ubiquitin Preservation Research
| Reagent | Function | Application Notes |
|---|---|---|
| N-Ethylmaleimide (NEM) | Irreversible cysteine protease DUB inhibitor | Use at 50-100 mM; Essential for K63 chain preservation |
| PR-619 | Broad-spectrum DUB inhibitor | Effective at 10-50 μM; Inhibits many cysteine DUBs |
| MG132 | Proteasome inhibitor | Use at 10-25 μM for 1-2 hours pre-lysis; Avoid extended treatment |
| TAK243 | E1 ubiquitin-activating enzyme inhibitor | Negative control; Depletes ubiquitin conjugates |
| EDTA/EGTA | Metalloprotease chelator | Inhibits JAMM/MPN+ metalloprotease DUBs; Use at 5-10 mM |
| Ubiquitin-Trap (Agarose/Magnetic) | Ubiquitin affinity resin | Pulls down diverse ubiquitinated proteins; Not linkage-specific |
| Linkage-specific ubiquitin antibodies | Detection of specific ubiquitin chain types | Variable quality between vendors; Requires validation |
Signaling Pathways and Experimental Workflows
Ubiquitin Preservation Pathway During Cell Lysis
Experimental Workflow for Ubiquitin Preservation
Establishing and consistently applying these ubiquitin preservation protocols is essential for generating reliable data in ubiquitination research. The key success factors include: (1) using comprehensive inhibitor cocktails at appropriate concentrations, (2) standardizing rapid processing techniques across all experiments, (3) implementing rigorous quality control measures, and (4) properly validating tools and reagents. By adopting these benchmarks, researchers can significantly improve the reproducibility and biological relevance of their findings in DUB inhibitor research and ubiquitin-related studies.
Conclusion
Effective prevention of deubiquitination during cell lysis through strategic DUB inhibitor implementation is fundamental to obtaining accurate, reproducible data in ubiquitin-proteasome system research. The integration of foundational DUB biology with optimized methodological approaches, systematic troubleshooting protocols, and rigorous validation techniques creates a comprehensive framework for preserving native ubiquitination states. As DUB-targeted therapies continue advancing toward clinical applications—with several inhibitors in preclinical development and early clinical trials—the importance of robust experimental techniques for assessing ubiquitination dynamics becomes increasingly critical. Future directions will likely see the development of more selective inhibitor cocktails, improved compatibility with emerging proteomic technologies, and standardized validation protocols across research communities. By implementing these evidence-based practices, researchers can significantly enhance experimental fidelity, accelerate drug discovery efforts, and advance our understanding of proteostasis in health and disease.
References
- 1. of selected deubiquitinating enzyme Effects on the... inhibitors [https://www.spandidos-publications.com/10.3892/ijo.2020.5034?text=fulltext]
- 2. of Inhibition by PR-619 induces apoptosis and... deubiquitination [https://pubmed.ncbi.nlm.nih.gov/33129231/]
- 3. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC2775171/]
- 4. Cell Lysate-Based AlphaLISA Deubiquitinase Assay Platform for ... [https://ncbi.nlm.nih.gov/pmc/articles/PMC5947317]
- 5. Reversible inactivation of deubiquitinases by... | Nature Communications [https://www.nature.com/articles/ncomms2532?error=cookies_not_supported]
- 6. Deubiquitinases (DUBs) and DUB inhibitors: a patent review - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4834700/]
- 7. - Wikipedia Deubiquitinating enzyme [https://en.wikipedia.org/wiki/Deubiquitinating_enzyme]
- 8. - Amerigo Scientific Deubiquitinating Enzyme Family [https://www.amerigoscientific.com/resource-deubiquitinating-enzyme-family.html]
- 9. Deubiquitinating enzymes (DUBs): Regulation, homeostasis, and oxidative stress response - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC8424594/]
- 10. Protein Partners of Deubiquitinating - PMC Enzymes [https://pmc.ncbi.nlm.nih.gov/articles/PMC2724835/]
- 11. Mechanisms for regulating deubiquitinating enzymes - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3970886/]
- 12. Cell Lysate-Based AlphaLISA Deubiquitinase Assay Platform for Identification of Small Molecule Inhibitors - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC5947317/]
- 13. Deubiquitinating enzymes: potential regulators of the tumor microenvironment and implications for immune evasion | Cell Communication and Signaling | Full Text [https://biosignaling.biomedcentral.com/articles/10.1186/s12964-024-01633-7]
- 14. Deubiquitinating enzymes and the proteasome regulate preferential sets of ubiquitin substrates | Nature Communications [https://www.nature.com/articles/s41467-022-30376-7]
- 15. Accelerating inhibitor discovery for deubiquitinating enzymes [https://www.nature.com/articles/s41467-023-36246-0?error=cookies_not_supported&code=88573156-5bbd-48f4-a8cf-9951cab872f6]
- 16. Recent advances in small molecule inhibitors of deubiquitinating enzymes - ScienceDirect [https://www.sciencedirect.com/science/article/abs/pii/S0223523425000893]
- 17. Cell Lysis and Fractionation Support—Troubleshooting | Thermo Fisher Scientific - US [https://www.thermofisher.com/us/en/home/technical-resources/technical-reference-library/protein-purification-isolation-support-center/cell-lysis-fractionation-support/cell-lysis-fractionation-support-troubleshooting.html]
- 18. Method for Measuring the Activity of Deubiquitinating Enzymes in Cell ... [https://www.jove.com/t/52784/method-for-measuring-activity-deubiquitinating-enzymes-cell-lines]
- 19. Deubiquitination and Activation of the NLRP3 Inflammasome by... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8552718/]
- 20. Advances in Deubiquitinating Enzyme Inhibition and Applications in Cancer Therapeutics - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7352412/]
- 21. Regulation of Wnt Signaling through Ubiquitination and Deubiquitination in Cancers - PubMed [https://pubmed.ncbi.nlm.nih.gov/32486158/]
- 22. Guide | Boster Bio Western Blot Troubleshooting [https://www.bosterbio.com/protocol-and-troubleshooting/western-blot-troubleshooting]
- 23. Systems for Profiling Deubiquitinating Activity - PMC Assay [https://pmc.ncbi.nlm.nih.gov/articles/PMC7460613/]
- 24. Recent advances in the development of deubiquitinases inhibitors as antitumor agents - ScienceDirect [https://www.sciencedirect.com/science/article/abs/pii/S0223523424000412]
- 25. Deubiquitinases in cancer | Oncotarget [https://www.oncotarget.com/article/3671/text/]
- 26. Accelerating inhibitor discovery for deubiquitinating enzymes | Nature Communications [https://www.nature.com/articles/s41467-023-36246-0]
- 27. The emerging role of deubiquitylating enzymes as therapeutic ... targets [https://pmc.ncbi.nlm.nih.gov/articles/PMC8935717/]
- 28. Deubiquitinating enzymes ( DUBs ): decipher underlying basis of... [https://pubmed.ncbi.nlm.nih.gov/34285347/]
- 29. Accelerating inhibitor discovery for... | Nature Communications [https://www.nature.com/articles/s41467-023-36246-0?error=cookies_not_supported]
- 30. PR-619 | DUB inhibitor [https://focusbiomolecules.com/pr-619-dub-inhibitor/]
- 31. A selective USP1–UAF1 inhibitor links deubiquitination to DNA... [https://www.nature.com/articles/nchembio.1455?error=cookies_not_supported]
- 32. Proteomics-Based Identification of DUB Substrates Using Selective Inhibitors - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7855594/]
- 33. Example Protocol: Cell Lysis for Downstream Protein Research | AAT Bioquest [https://www.aatbio.com/resources/application-notes/example-protocol-cell-lysis-for-downstream-protein-research]
- 34. Identification of deubiquitinase inhibitors via high-throughput ... [https://ncbi.nlm.nih.gov/pmc/articles/PMC8536791]
- 35. Frontiers | Recent Developments in Cell Permeable Deubiquitinating Enzyme Activity-Based Probes [https://www.frontiersin.org/journals/chemistry/articles/10.3389/fchem.2019.00876/full]
- 36. Method for Measuring the Activity of Deubiquitinating Enzymes in Cell Lines and Tissue Samples - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4542631/]
- 37. Deubiquitinase inhibitor - PR reduces... | PLOS One 619 [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0202409]
- 38. The broad-spectrum deubiquitinating enzyme inhibitor - PR ... 619 [https://www.nature.com/articles/s41598-024-75562-3?error=cookies_not_supported&code=1d49adb5-6608-4ecf-9e79-79fd93e10a51]
- 39. Cell Lysis | Thermo Fisher Scientific - RU Buffers [https://www.thermofisher.com/ru/ru/home/life-science/protein-biology/protein-purification-isolation/cell-lysis-fractionation/cell-lysis-total-protein-extraction.html]
- 40. Lysis buffer - Wikipedia [https://en.wikipedia.org/wiki/Lysis_buffer]
- 41. solubilization of TRIzol-precipitated protein permits Western... Optimized [https://pmc.ncbi.nlm.nih.gov/articles/PMC5392113/]
- 42. Choosing The Right Lysis Buffer | Proteintech Group [https://www.ptglab.com/support/western-blot-protocol/choosing-the-right-lysis-buffer/]
- 43. Why is my lysis buffer not working? 9 Lysis Buffer Issues – GoldBio [https://www.goldbio.com/blogs/articles/why-is-my-lysis-buffer-not-working-how-to-resolve-9-lysis-buffer-issues-that-might-be-holding-you-back]
- 44. Lysate preparation protocol for western blotting | Abcam [https://www.abcam.com/en-us/technical-resources/protocols/lysate-preparation-for-western-blot]
- 45. Sample preparation for western blot | Abcam [https://www.abcam.com/en-us/technical-resources/guides/western-blot-guide/sample-preparation-for-western-blot]
- 46. multi-omics data reveals function and therapeutic potential... Integrating [https://elifesciences.org/articles/72879]
- 47. Detection of Protein Ubiquitination - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3149903/]
- 48. Tandem Affinity : Principles, Purification , and Applications Techniques [https://www.creative-proteomics.com/resource/tandem-affinity-purification-principles-techniques-applications.htm]
- 49. Protein Immunoprecipitation (IP), Co-Immunoprecipitation ( Co - IP ), and... [https://www.thermofisher.com/in/en/home/technical-resources/technical-reference-library/protein-purification-isolation-support-center/protein-ip-coip-pulldown-support/protein-ip-coip-pulldown-support-troubleshooting.html]
- 50. A Post-translational Modification–enhanced Pull - down Method to... [https://bio-protocol.org/en/bpdetail?id=4816&type=0]
- 51. Target-Class Selectivity Profiling for Deubiquitinase ( DUB ) Inhibitors [https://www.technologynetworks.com/drug-discovery/application-notes/target-class-selectivity-profiling-for-deubiquitinase-dub-inhibitors-356522]
- 52. cOmplete™ Lysis-M Protocol & Troubleshooting [https://www.sigmaaldrich.com/US/en/technical-documents/technical-article/protein-biology/protein-lysis-and-extraction/complete-lysis-m]
- 53. The proteasome deubiquitinase inhibitor VLX1570 shows selectivity ... [https://www.nature.com/articles/srep26979?error=cookies_not_supported]
- 54. Lysis of Mammalian and Sf9 Cells - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4352304/]
- 55. -site-centric chemoproteomics... | Nature Communications Reactive [https://www.nature.com/articles/s41467-018-03511-6?error=cookies_not_supported&code=0354f5b9-4526-40a5-b335-18571330367b]
- 56. Interplay between bacterial deubiquitinase and ubiquitin E3 ligase... [https://elifesciences.org/articles/58114]
- 57. Protein purification and analysis: next generation Western blotting techniques - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6810642/]
- 58. | Thermo Fisher Scientific - TR Cell Lysis Buffers [https://www.thermofisher.com/tr/en/home/life-science/protein-biology/protein-purification-isolation/cell-lysis-fractionation/cell-lysis-total-protein-extraction.html]
- 59. Methods: A Guide to Cell Protein Extraction | Boster Bio Lysis Efficient [https://www.bosterbio.com/blog/post/cell-lysis-the-first-and-crucial-step-in-molecular-biology]
- 60. Lysate Preparation - RIPA Cell Method | Fortis Life Sciences Buffer [https://www.fortislife.com/protocols/antibody-protocols/cell-lysate-preparation-ripa-method]
- 61. Development of a simple method for protein conjugation by copper-free click reaction and its application to antibody-free Western blot analysis - PubMed [https://pubmed.ncbi.nlm.nih.gov/23039792/]
- 62. Deubiquitinating enzymes as oncotargets | Oncotarget [https://www.oncotarget.com/article/3922/text/]
- 63. Emerging Role of Deubiquitinating Enzymes (DUBs) in Melanoma... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9322030/]
- 64. The emerging role of deubiquitylating enzymes as therapeutic targets in cancer metabolism | Cancer Cell International | Full Text [https://cancerci.biomedcentral.com/articles/10.1186/s12935-022-02524-y]
- 65. A Review on Macroscale and Microscale Cell Lysis Methods - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6190294/]
- 66. Traditional Methods of Cell Lysis | Thermo Fisher Scientific - US [https://www.thermofisher.com/us/en/home/life-science/protein-biology/protein-biology-learning-center/protein-biology-resource-library/pierce-protein-methods/traditional-methods-cell-lysis.html]
- 67. EOAI, a ubiquitin-specific peptidase 5 inhibitor , prevents non-small... [https://bmccancer.biomedcentral.com/articles/10.1186/s12885-023-10506-0]
- 68. Re-Evaluating the Mechanism of α, β-Unsaturated Carbonyl... of Action [https://pubmed.ncbi.nlm.nih.gov/32109059/]
- 69. The optimization of in vitro high-throughput chemical lysis of Escherichia coli. Application to ACP domain of the polyketide synthase ppsC from Mycobacterium tuberculosis - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC2855807/]
- 70. – Western | Thermo Fisher Scientific - ZA Blotting Troubleshooting [https://www.thermofisher.com/za/en/home/technical-resources/technical-reference-library/protein-electrophoresis-western-blotting-support-center/western-blotting-support/western-blotting-support-troubleshooting.html]
- 71. Guide - TotalLab Western Blot Troubleshooting [https://totallab.com/resources/western-blot-troubleshooting-guide/]
- 72. Recent Advances in the Discovery of Deubiquitinating Enzyme Inhibitors - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7112275/]
- 73. as a tool for Functional Proteomic Analysis of... Activity Based Probes [https://pmc.ncbi.nlm.nih.gov/articles/PMC2997944/]
- 74. Sample Quantitation Support— Mass Spectrometry Troubleshooting [https://www.thermofisher.com/nz/en/home/technical-resources/technical-reference-library/mass-spectrometry-support-center/mass-spectrometry-reagents-support/ms-sample-quantitation-support/ms-sample-quantitation-support-troubleshooting.html]
- 75. Mastering Mass Techniques Spectrometry [https://www.numberanalytics.com/blog/mastering-mass-spectrometry-techniques-instrumental-analysis]
- 76. ( Mass ) Spectrometer guide | Alliance Bioversity... MS troubleshooting [https://alliancebioversityciat.org/publications-data/mass-spectrometer-ms-troubleshooting-guide]
- 77. Biochemistry, Ubiquitination - StatPearls - NCBI Bookshelf [https://www.ncbi.nlm.nih.gov/books/NBK556052/]
- 78. Measuring Enzymatic Activity of Neurodevelopmental... | JoVE [https://www.jove.com/it/t/67367/measuring-enzymatic-activity-neurodevelopmental-disorder-associated]
- 79. Suppression of the Deubiquitinating Enzyme USP5 Causes the... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2696100/]
- 80. Disrupting USP39 deubiquitinase function impairs the survival and... [https://jeccr.biomedcentral.com/articles/10.1186/s13046-024-03241-2]
- 81. The Length of a Ubiquitin Chain: A General Factor for Selective Recognition by Ubiquitin-Binding Proteins - PubMed [https://pubmed.ncbi.nlm.nih.gov/32301549/]
- 82. Methods to measure ubiquitin chain length and linkage - PubMed [https://pubmed.ncbi.nlm.nih.gov/30850048/]
- 83. Branched ubiquitin binding and deubiquitination by UCH37... chain [https://elifesciences.org/articles/72798]
- 84. Assessment of Ubiquitin Chain Topology by Targeted Mass Spectrometry - PubMed [https://pubmed.ncbi.nlm.nih.gov/30980320/]
- 85. Clinically used antirheumatic agent auranofin is... | Oncotarget [https://www.oncotarget.com/article/2113/text/]
- 86. Frontiers | ABPP-HT - High-Throughput Activity-Based Profiling of... [https://www.frontiersin.org/journals/chemistry/articles/10.3389/fchem.2021.640105/full]
- 87. Protease Inhibitor Cocktail (EDTA-Free, 100× in DMSO) | TargetMol [https://www.targetmol.com/reagent/protease%20inhibitor%20cocktail]
- 88. UL36 Encoded by Marek’s Disease Virus Exhibits Linkage-Specific... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7084888/]
- 89. Identification and validation of Selective Deubiquitinase Inhibitors - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9473745/]
- 90. - PR | Cell Signaling Technology 619 [https://www.cellsignal.com/products/activators-inhibitors/pr-619/26065]
- 91. The Deubiquitinating Enzyme Inhibitor - PR is a Potent DNA... 619 [https://pubmed.ncbi.nlm.nih.gov/31515282/]
- 92. How to Better Study Ubiquitination | Proteintech Group [https://www.ptglab.com/news/blog/how-to-better-study-ubiquitination/]
- 93. Deubiquitinating enzymes and the... | Nature Communications [https://www.nature.com/articles/s41467-022-30376-7?error=cookies_not_supported&code=61f8a004-e32c-47e0-9c1b-671d0697a587]
- 94. Frontiers | The emerging role and therapeutic implications of bacterial... [https://www.frontiersin.org/articles/10.3389/fimmu.2023.1303072/full?field=&journalName=Frontiers_in_Immunology&id=1303072]
- 95. Deubiquitylating enzymes and drug discovery: emerging opportunities | Nature Reviews Drug Discovery [https://www.nature.com/articles/nrd.2017.152]
- 96. Extracting Better Ubiquitin Data from your Samples [https://bitesizebio.com/31971/extracting-better-ubiquitin-data-samples/]