Decoding the Ubiquitinome: A Comprehensive Guide to UBD-Based Affinity Enrichment Methods
This article provides a comprehensive resource for researchers on Ubiquitin-Binding Domain (UBD)-based affinity enrichment, a cornerstone technique for profiling the ubiquitinome.
Decoding the Ubiquitinome: A Comprehensive Guide to UBD-Based Affinity Enrichment Methods
Abstract
This article provides a comprehensive resource for researchers on Ubiquitin-Binding Domain (UBD)-based affinity enrichment, a cornerstone technique for profiling the ubiquitinome. It covers the foundational principles of ubiquitin biology and the diversity of UBDs, then details established and emerging methodological protocols, including TUBEs, FUBEs, and linkage-specific tools. The guide offers practical troubleshooting and optimization strategies for common experimental challenges and presents a comparative analysis of method validation. Aimed at scientists and drug development professionals, this review synthesizes current knowledge to enable robust experimental design and advance the study of ubiquitin signaling in health and disease.
Understanding the Ubiquitin Code: The Foundation of UBD-Based Enrichment
Ubiquitination is a crucial post-translational modification that regulates nearly all cellular processes in eukaryotic cells, ranging from targeted protein degradation via the proteasome to DNA repair, cell cycle regulation, and immune responses [1] [2]. This modification involves the covalent attachment of ubiquitin, an 8 kDa protein, to target substrates through a sequential enzymatic cascade involving E1 (activating), E2 (conjugating), and E3 (ligase) enzymes [2]. The complexity of ubiquitin signaling arises from the ability of ubiquitin itself to form polymers through its seven lysine residues (K6, K11, K27, K29, K33, K48, K63) and N-terminal methionine (M1), creating diverse polyubiquitin chains with distinct biological functions [3]. For instance, K48-linked chains primarily target proteins for proteasomal degradation, while K63-linked chains regulate signal transduction and protein trafficking [2].
The study of ubiquitination has been revolutionized by affinity enrichment methods utilizing ubiquitin-binding domains (UBDs), which are protein modules that recognize and bind to ubiquitin modifications [4] [1]. These tools enable researchers to capture and analyze ubiquitinated proteins from complex biological samples, addressing the challenges posed by the low abundance of ubiquitinated species, the transient nature of ubiquitination, and the activity of deubiquitinating enzymes (DUBs) [5]. Recent advancements in UBD engineering have yielded reagents with enhanced affinity, specificity, and reduced linkage bias, significantly improving our ability to decipher the ubiquitin code under physiological and pathological conditions [4] [1] [5].
Advanced UBD-Based Tools for Ubiquitin Enrichment
Key UBD Technologies and Their Characteristics
The current landscape of UBD-based affinity tools features several engineered domains with distinct properties and applications. These tools can be broadly categorized into pan-specific UBDs that recognize all ubiquitin chain types, and linkage-specific UBDs that selectively bind particular ubiquitin chain architectures.
Table 1: Comparison of Key UBD-Based Affinity Tools
| UBD Tool | Source/Design | Affinity & Specificity | Key Applications | Advantages |
|---|---|---|---|---|
| OtUBD | Derived from Orientia tsutsugamushi | High-affinity for mono- and polyubiquitin; pan-specific | Immunoblotting, proteomics, UbiCREST | Versatile, economical, works with various lysate types [4] |
| ThUBD | Tandem hybrid UBD | High-affinity, unbiased toward different ubiquitin chains | High-throughput screening (96-well plates), TUF-WB, deep ubiquitinome profiling | No linkage bias, high sensitivity, suitable for low-input samples [1] [5] |
| TUBEs (Tandem Ubiquitin Binding Entities) | Multiple ubiquitin-binding domains in tandem | Nanomolar affinity; available in pan- and chain-specific variants | Studying linkage-specific functions, PROTAC development | Protection from DUBs, chain-specific versions available [2] |
| K63-TUBEs | Engineered TUBE variant | Specific for K63-linked chains | Analysis of inflammatory signaling pathways | Enables study of non-degradative ubiquitination in signal transduction [2] |
| K48-TUBEs | Engineered TUBE variant | Specific for K48-linked chains | Monitoring PROTAC-induced degradation | Specific detection of degradative ubiquitination [2] |
Quantitative Performance of UBD-Based Methods
The effectiveness of UBD-based methods can be evaluated through specific quantitative parameters that measure enrichment efficiency, sensitivity, and specificity.
Table 2: Quantitative Performance Metrics of UBD-Based Enrichment Methods
| Method | Enrichment Efficiency | Detection Sensitivity | Reproducibility | Special Features |
|---|---|---|---|---|
| OtUBD Protocol | Strong enrichment of mono- and polyubiquitinated proteins | Compatible with immunoblotting and LC-MS/MS | High with optimized buffer formulations | Native and denaturing workflows available [4] |
| ThUBD 96-well Plate | Binds ~5 pmol of polyubiquitin chains | High-throughput, rapid detection | High inter-assay consistency | Unbiased recognition of all ubiquitin chain types [1] |
| DRUSP with ThUBD | ~10-fold improvement in ubiquitin signal enrichment | Extracts more ubiquitinated proteins | Enhanced stability and reproducibility | Works with denatured samples, refolding step included [5] |
| Chain-specific TUBEs | Faithful capture of linkage-specific ubiquitination | Enables monitoring endogenous target protein ubiquitination | Context-dependent linkage specificity | Differentiates K48 vs K63 ubiquitination in cellular contexts [2] |
Detailed Experimental Protocols
OtUBD-Based Affinity Enrichment for Ubiquitinated Proteins
This protocol describes a step-by-step process for enriching ubiquitinated proteins from baker's yeast and mammalian cell lysates using OtUBD, which can strongly enrich both mono- and polyubiquitinated proteins from crude lysates [4].
Reagents and Equipment
- OtUBD affinity resin (prepared by coupling recombinant OtUBD to appropriate chromatography matrix)
- Lysis buffer (native): 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% NP-40, supplemented with protease inhibitors and DUB inhibitors (e.g., 10 mM N-ethylmaleimide)
- Denaturing lysis buffer: 6 M urea or 1% SDS in PBS with DTT and protease inhibitors
- Wash buffer 1: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.1% NP-40
- Wash buffer 2: 50 mM Tris-HCl (pH 7.5), 500 mM NaCl, 1 mM EDTA, 0.1% NP-40
- Elution buffer: 100 mM glycine (pH 2.5) or SDS-PAGE sample buffer for direct analysis
- Equipment: Centrifuge, end-over-end rotator, chromatography columns or spin filters, electrophoresis system
Procedure
Sample Preparation:
- For native conditions: Harvest cells and lyse in native lysis buffer (1 mL per 10^7 cells) by gentle vortexing. Incubate on ice for 30 minutes with occasional mixing. Clarify by centrifugation at 16,000 × g for 15 minutes at 4°C.
- For denaturing conditions: Lyse cells directly in denaturing lysis buffer. Heat at 95°C for 5 minutes to denature proteins and inhibit DUBs. Dilute 10-fold with PBS before proceeding to step 2.
Affinity Pulldown:
- Equilibrate OtUBD affinity resin with appropriate lysis buffer.
- Incubate clarified lysate with OtUBD resin (50 μL bed volume per 1 mg total protein) for 2 hours at 4°C with end-over-end mixing.
- Centrifuge at 1,000 × g for 2 minutes and carefully remove supernatant.
Washing:
- Wash resin sequentially with 10 bed volumes of wash buffer 1, followed by 10 bed volumes of wash buffer 2.
- Perform a final wash with 10 bed volumes of PBS or appropriate assay buffer.
Elution:
- Elute bound proteins with 2-3 bed volumes of elution buffer by incubating for 5 minutes at room temperature with gentle mixing.
- Neutralize acidic eluates immediately with 1 M Tris-HCl (pH 8.0).
- Analyze eluates by immunoblotting or mass spectrometry.
Controls:
- Include control resin without coupled OtUBD to assess non-specific binding.
- Use known ubiquitinated proteins as positive controls when available.
DRUSP (Denatured-Refolded Ubiquitinated Sample Preparation) with ThUBD
The DRUSP method addresses limitations of native lysis conditions by implementing a denaturation-refolding strategy that significantly improves ubiquitinated protein recovery and reproducibility [5].
Reagents and Equipment
- Strong denaturation buffer: 8 M urea, 2% SDS, 50 mM Tris-HCl (pH 7.5), 5 mM DTT
- Refolding buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.1% NP-40, 10% glycerol
- ThUBD affinity resin (prepared as described in [1] [5])
- Ultrafiltration devices (10 kDa MWCO)
- DUB inhibitor cocktail
Procedure
Denaturing Extraction:
- Lyse cells or tissue directly in strong denaturation buffer (1 mL per 10^7 cells).
- Heat at 95°C for 10 minutes with vigorous shaking to ensure complete denaturation and inactivation of DUBs.
- Cool to room temperature and dilute with 4 volumes of refolding buffer.
Refolding:
- Concentrate the diluted lysate using ultrafiltration devices (10 kDa MWCO) according to manufacturer's instructions.
- Simultaneously exchange buffer to refolding buffer during concentration.
- Adjust final protein concentration to 1-2 mg/mL with refolding buffer.
ThUBD Enrichment:
- Incubate refolded lysate with ThUBD affinity resin (50 μL bed volume per 1 mg total protein) for 2 hours at 4°C.
- Wash resin with 10 bed volumes of refolding buffer containing 0.1% NP-40.
- Elute with SDS-PAGE sample buffer or compatible elution buffer for downstream applications.
Quality Assessment:
- Monitor ubiquitin signal enrichment by immunoblotting with anti-ubiquitin antibodies.
- Assess specificity by comparing to control samples without ThUBD resin.
UBD Applications in Drug Discovery and PROTAC Development
The development of Proteolysis Targeting Chimeras (PROTACs) has emerged as a promising therapeutic strategy that hijacks the ubiquitin-proteasome system to target specific proteins for degradation [1] [2]. UBD-based tools play a critical role in characterizing PROTAC efficiency and mechanism of action by enabling monitoring of target protein ubiquitination.
Chain-specific TUBEs have been successfully applied to investigate the ubiquitination dynamics of Receptor-Interacting Serine/Threonine-Protein Kinase 2 (RIPK2), a key regulator of inflammatory signaling [2]. In this application, K63-TUBEs specifically captured L18-MDP-stimulated K63 ubiquitination of RIPK2, while K48-TUBEs detected RIPK2 PROTAC-induced K48 ubiquitination. This approach demonstrates how chain-selective UBDs can differentiate context-dependent ubiquitin linkages, providing crucial information for drug development.
High-throughput screening platforms utilizing ThUBD-coated 96-well plates enable rapid assessment of compound libraries for their effects on protein ubiquitination [1]. These assays overcome limitations of traditional Western blotting, which is low-throughput and provides only semiquantitative data. The optimized ThUBD plate assay allows specific binding to approximately 5 pmol of polyubiquitin chains, enabling sensitive detection of ubiquitination changes in response to candidate therapeutics.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagent Solutions for UBD-Based Ubiquitin Enrichment
| Reagent | Function & Application | Examples/Specifications |
|---|---|---|
| OtUBD Affinity Resin | Enrichment of mono- and polyubiquitinated proteins from various lysates | Compatible with native and denaturing conditions; works with yeast and mammalian cells [4] |
| ThUBD-Coated Plates | High-throughput detection of ubiquitination signals in 96-well format | Corning 3603-type plates coated with 1.03 μg ± 0.002 of ThUBD [1] |
| Chain-Specific TUBEs | Selective capture of linkage-specific polyubiquitination | K48-TUBEs and K63-TUBEs with nanomolar affinities [2] |
| DUB Inhibitor Cocktails | Preservation of ubiquitin signals during sample preparation | Include N-ethylmaleimide, PR-619, or specific DUB inhibitors [4] [5] |
| Ubiquitin Chain Standards | Quality control and assay standardization | Recombinant ubiquitin chains of defined linkages (K48, K63, etc.) [1] |
| DRUSP Buffer Systems | Denaturing extraction with subsequent refolding for improved ubiquitin recovery | 8 M urea, 2% SDS, with refolding through ultrafiltration [5] |
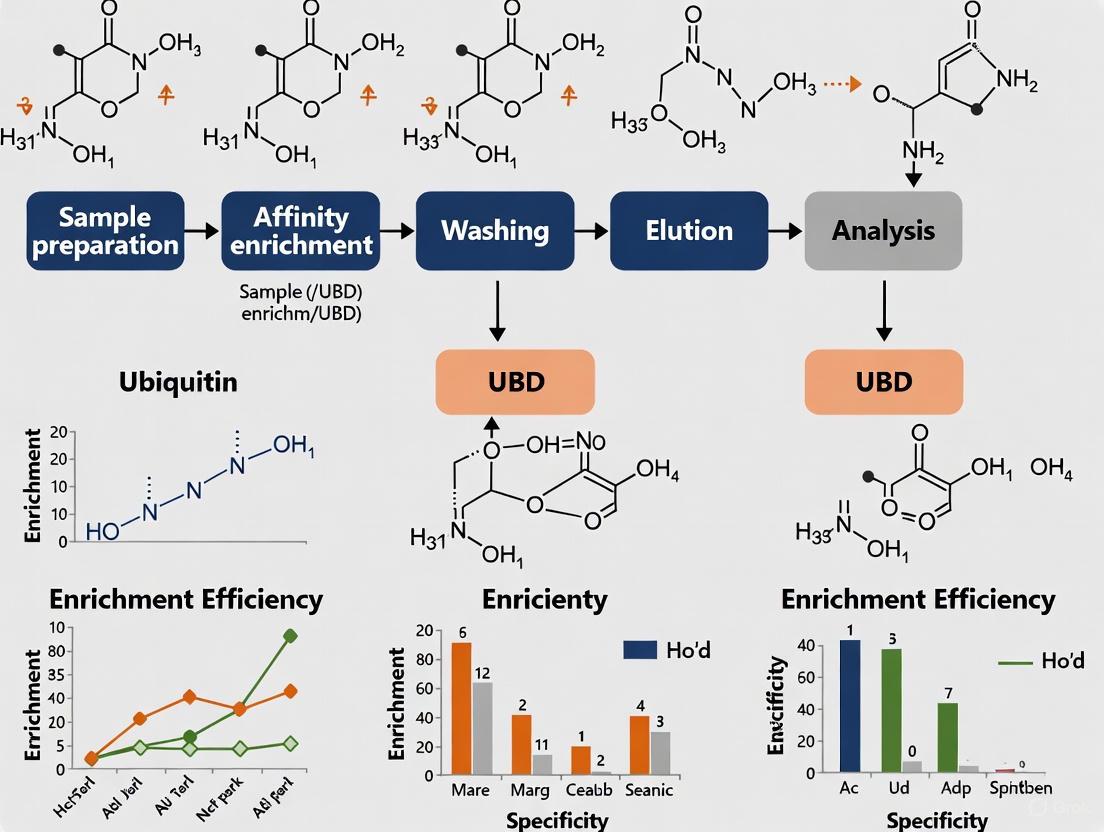
Visualizing Experimental Workflows and Ubiquitin Signaling
The following diagrams illustrate key experimental workflows and ubiquitin signaling pathways relevant to UBD-based affinity enrichment methods.
UBD-Based Ubiquitin Enrichment Workflow
Ubiquitin Chain Linkage Recognition by UBDs
PROTAC-Induced Ubiquitination Detection
UBD-based affinity enrichment methods have revolutionized our ability to study the complex landscape of ubiquitin modifications. The continuous development of engineered UBDs with enhanced affinity, reduced linkage bias, and specialized applications has provided researchers with powerful tools to decipher ubiquitin signaling in health and disease. These methodologies now enable highly specific detection of ubiquitination events, support high-throughput drug discovery efforts, and facilitate the characterization of novel therapeutic modalities such as PROTACs. As these tools continue to evolve, they will undoubtedly yield further insights into the intricate workings of the ubiquitin-proteasome system and its applications in biomedical research and therapeutic development.
Ubiquitin-binding domains (UBDs) are modular protein elements that serve as critical decoders of the ubiquitin code, a pervasive post-translational regulatory system in eukaryotic cells. These domains recognize and bind non-covalently to ubiquitin signals, translating them into specific cellular outcomes such as protein degradation, DNA repair, and immune signaling [6]. The integration of UBDs into affinity enrichment tools has revolutionized the study of ubiquitination, enabling researchers to capture and analyze ubiquitinated substrates with high specificity and affinity. This application note details the principles, protocols, and key reagent solutions for UBD-based methodologies, providing a framework for their application in basic research and drug discovery.
Ubiquitination involves the covalent attachment of the small protein ubiquitin to substrate proteins, fundamentally altering their fate, function, or localization. The versatility of this signal arises from the capacity of ubiquitin itself to form polymers (polyubiquitin chains) through any of its seven lysine residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, Lys63) or its N-terminus (linear chains), with different linkage types encoding distinct functional consequences [6]. For instance, Lys48-linked chains typically target substrates for proteasomal degradation, whereas Lys63-linked and linear chains are more often involved in signaling pathways such as NF-κB activation [6].
Ubiquitin-binding domains (UBDs) are the readers of this complex language. It is estimated that the human proteome contains more than 150 proteins harboring UBDs, which are classified into approximately 20 different families based on their structure [6] [7]. These domains are typically small (often less than 50 amino acids) and exhibit a remarkable diversity of folds, including α-helical structures, zinc fingers, and pleckstrin homology (PH) domains [7]. Despite their structural variation, many UBDs bind to a common hydrophobic surface patch on ubiquitin centered around the Ile44 residue [6] [7]. A key characteristic of most UBDs is their relatively weak affinity (in the low to mid-μM range) for monoubiquitin, which allows for reversible and dynamic interactions in vivo [7]. Specificity is often achieved through avidity effects, whereby a protein with multiple UBDs synergistically binds to multiple ubiquitin subunits in a polyubiquitin chain [6]. Furthermore, some UBDs exhibit linkage specificity, preferentially binding one type of polyubiquitin chain over another, which is crucial for directing specific cellular responses [6] [8].
UBD-Based Research Reagent Solutions
The exploitation of UBDs in biochemical tools has overcome historical limitations of antibody-based methods, such as low affinity and linkage bias. The table below summarizes key reagent solutions used in the field.
Table 1: Key Research Reagent Solutions for UBD-Based Ubiquitin Enrichment
| Reagent / Tool | Description | Key Features and Applications |
|---|---|---|
| OtUBD | A high-affinity UBD derived from Orientia tsutsugamushi [4]. | - Strong enrichment of mono- and poly-ubiquitinated proteins from crude lysates.- Used in native (for ubiquitin interactome) or denaturing (for covalent ubiquitinome) workflows.- Compatible with immunoblotting and LC-MS/MS proteomics [4]. |
| ThUBD | An engineered tandem hybrid ubiquitin-binding domain [1]. | - Exhibits high affinity for polyubiquitinated proteins and no bias towards different ubiquitin chain types.- Used in assays like TUF-WB and coated onto 96-well plates for high-throughput, sensitive detection of ubiquitination signals [1]. |
| TUBE (Tandem Ubiquitin Binding Entity) | A recombinant tool containing multiple ubiquitin-binding domains in tandem. | - Increases avidity for polyubiquitin chains.- Used in PROTAC assay plates for high-throughput screening.- Can exhibit some linkage bias and lower affinity compared to newer tools [1]. |
| ThUBD-coated 96-well plates | High-density plates coated with ThUBD for capture assays [1]. | - Enables high-throughput, unbiased quantification of proteins modified by all ubiquitin chain types.- A single well can specifically bind ~5 pmol of polyubiquitin chains [1]. |
| NZF Domains | A family of compact zinc finger UBDs (e.g., in TAB2, HOIP) [8]. | - Some exhibit linkage specificity (e.g., TAB2 for Lys63 and phosphorylated Lys6 chains).- Can utilize secondary interaction surfaces to bind specifically to ubiquitinated substrates like NEMO, adding a layer of specificity [8]. |
The functional characteristics of UBDs and the performance metrics of tools derived from them are critical for experimental design. The following table consolidates key quantitative data.
Table 2: Performance Metrics of UBDs and Associated Tools
| Parameter | Typical Range or Value | Context and Notes |
|---|---|---|
| Number of Human UBD Families | ~20 - 29 types [6] [7] | Constant discovery of new UBDs expands the ubiquitin interactome. |
| Binding Affinity (Monoubiquitin) | Low to mid μM range [7] | Weak affinity allows for reversible signaling; avidity enhances effective affinity for chains. |
| ThUBD Coating Capacity | ~5 pmol of polyubiquitin chains per well [1] | Refers to the binding capacity of a single well in a 96-well plate coated with 1.03 μg of ThUBD. |
| Key Ubiquitin Surface Patches | Ile44 patch, Ile36 patch, C-terminal diglycine motif [7] | Different UBDs bind to distinct surface patches on ubiquitin to achieve functional diversity. |
| WCAG Contrast Ratio for Visuals | 4.5:1 (normal text), 3:1 (large text) [9] [10] | Critical guideline for ensuring accessibility in generated diagrams and figures for publications and presentations. |
Detailed Experimental Protocols
Protocol: Enrichment of Ubiquitinated Proteins using OtUBD Affinity Resin
This protocol describes the use of the high-affinity OtUBD to enrich ubiquitinated proteins from yeast or mammalian cell lysates, allowing for subsequent analysis by immunoblotting or mass spectrometry [4].
Key Materials:
- OtUBD affinity resin
- Lysis Buffer (e.g., Native or Denaturing, depending on the goal)
- Wash Buffer
- Elution Buffer (e.g., containing SDS or competing free ubiquitin)
- Protease and Deubiquitinase Inhibitors
Methodology:
Cell Lysis and Sample Preparation:
- Harvest cells and lyse them using an appropriate buffer. The choice of buffer is crucial:
- Native Lysis Buffer: Use a non-denaturing buffer (e.g., containing Tris-HCl, NaCl, and NP-40) to preserve non-covalent protein-protein interactions. This workflow will co-purify both covalently ubiquitinated proteins and their non-covalent interacting partners (the "ubiquitin interactome") [4].
- Denaturing Lysis Buffer: Use a buffer containing strong denaturants (e.g., SDS, urea) to disrupt all non-covalent interactions. This workflow isolates only the covalently ubiquitinated proteins (the "ubiquitinome") [4].
- Add protease and deubiquitinase inhibitors to the lysis buffer to prevent degradation of ubiquitin conjugates during processing.
- Clarify the lysate by centrifugation at high speed (e.g., 16,000 × g for 15 minutes) to remove insoluble debris.
- Harvest cells and lyse them using an appropriate buffer. The choice of buffer is crucial:
Affinity Purification (Pulldown):
- Incubate the clarified cell lysate with the OtUBD affinity resin for 1-2 hours at 4°C with gentle agitation.
- Pellet the resin by brief centrifugation and carefully remove the supernatant.
Washing:
- Wash the resin multiple times (typically 3-5 times) with a wash buffer compatible with the lysis conditions. The wash buffer should contain a moderate salt concentration (e.g., 150-300 mM NaCl) to reduce non-specific binding without eluting genuine ubiquitin conjugates.
Elution:
- Elute the bound ubiquitinated proteins from the resin. Two common methods are:
- SDS Elution: Directly add 1X SDS-PAGE loading buffer to the resin and heat at 95°C for 5-10 minutes. This is the most common and effective method for downstream immunoblotting.
- Competitive Elution: Incubate the resin with a buffer containing a high concentration of free ubiquitin (e.g., 1-2 mg/mL) to compete for binding to OtUBD. This native elution is gentler and can be preferable for certain functional studies or specific proteomic applications [4].
- Elute the bound ubiquitinated proteins from the resin. Two common methods are:
Downstream Analysis:
- Analyze the eluates by immunoblotting with antibodies against ubiquitin or proteins of interest.
- For proteomic analysis, the eluates can be subjected to tryptic digestion and Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) to identify the ubiquitinated proteins and, if using specific diglycine remnant enrichment antibodies, the specific modification sites [4].
Diagram 1: OtUBD affinity enrichment workflow.
Protocol: High-Throughput Detection of Ubiquitination using ThUBD-Coated Plates
This protocol leverages ThUBD-coated 96-well plates for the sensitive, unbiased, and high-throughput quantification of ubiquitination signals from complex proteome samples [1].
Key Materials:
- ThUBD-coated high-density 96-well plates (e.g., Corning 3603)
- Recombinant ThUBD protein
- Blocking Buffer (e.g., 5% BSA in TBST)
- Assay Buffer
- Detection Antibody (e.g., ThUBD-HRP conjugate or primary anti-ubiquitin antibody with HRP-secondary)
- Chemiluminescent or Colorimetric Substrate
Methodology:
Plate Preparation:
- The plates are pre-coated with ThUBD (optimally 1.03 μg ± 0.002 per well) [1].
- Block the plates with a suitable blocking buffer (e.g., 5% BSA) for 1 hour at room temperature to prevent non-specific binding.
- Wash the plates 2-3 times with an assay buffer or wash buffer.
Sample Incubation and Capture:
- Add prepared cell lysates (in a compatible, non-denaturing buffer) or purified protein samples to the wells.
- Incubate for 1-2 hours at room temperature with gentle shaking to allow ubiquitinated proteins to bind to the immobilized ThUBD.
Washing:
- Thoroughly wash the wells multiple times (e.g., 5 times) with a wash buffer to remove unbound proteins and contaminants.
Detection:
- For direct detection, add a ThUBD-HRP conjugate to bind the captured ubiquitinated proteins, creating a sensitive sandwich assay [1].
- Alternatively, add a primary anti-ubiquitin antibody, followed by an HRP-conjugated secondary antibody.
- After incubation and subsequent washing, develop the signal by adding a chemiluminescent or colorimetric substrate.
- Measure the signal using a plate reader.
Data Analysis:
- Quantify the ubiquitination signal relative to standards or controls. This platform allows for the screening of multiple conditions in parallel, making it ideal for drug discovery applications, such as evaluating the efficacy of PROTACs or DUB inhibitors [1].
Applications in Research and Drug Discovery
UBD-based tools are indispensable in modern biological research and therapeutic development. Their primary applications include:
- Target Deconvolution for PROTACs: Proteolysis Targeting Chimeras (PROTACs) are heterobifunctional molecules that induce targeted protein degradation. UBD-based assays, like those using ThUBD-coated plates, are used to confirm and quantify the induced ubiquitination of target proteins, a critical step in the PROTAC mechanism of action [1].
- Ubiquitinome Profiling: By combining OtUBD or ThUBD enrichment with LC-MS/MS, researchers can perform system-wide profiling of ubiquitinated proteins under different physiological or pathological conditions, identifying novel substrates and dynamics in the ubiquitin code [4] [1].
- Studying Linkage-Specific Signaling: The use of UBDs with known linkage preferences (e.g., specific NZF domains) allows researchers to dissect the roles of particular chain types in pathways like NF-κB signaling and mitophagy [6] [8].
- Diagnostic and Prognostic Assay Development: The high sensitivity and throughput of tools like ThUBD-coated plates hold potential for developing clinical assays to monitor disease-associated ubiquitination signatures in patient samples.
Troubleshooting Guide
Table 3: Common Issues and Solutions in UBD-Based Assays
| Problem | Potential Cause | Suggested Solution |
|---|---|---|
| High Background Noise | Incomplete blocking or non-optimal washing. | Increase blocking time; optimize wash buffer stringency (e.g., increase salt concentration, add mild detergents); include more wash cycles. |
| Low Signal/Weak Enrichment | Insufficient lysis; degradation of conjugates; low affinity of UBD. | Ensure fresh deubiquitinase inhibitors are used; verify lysis efficiency; increase input protein amount; consider using a higher-affinity UBD like OtUBD or ThUBD. |
| Bias in Ubiquitin Chain Detection | Use of a UBD with inherent linkage preference (e.g., some TUBEs). | Employ an unbiased UBD tool such as ThUBD for a comprehensive view of all ubiquitination types [1]. |
| Inconsistent Results in HTS | Plate coating inconsistency or evaporation in edge wells. | Use quality-controlled, pre-coated plates; include controls in all plates; use plate seals during incubation steps. |
Ubiquitin-binding domains (UBDs) are modular elements within effector proteins that recognize ubiquitin (Ub) signals non-covalently, serving as critical interpreters of the ubiquitin code [6]. The versatility of ubiquitin signaling stems from the capacity of ubiquitin itself to form diverse polymeric chains. Through an enzymatic cascade involving E1, E2, and E3 enzymes, the C-terminus of one ubiquitin can be attached to any of seven lysine residues (K6, K11, K27, K29, K33, K48, K63) or the N-terminal methionine (M1) of another ubiquitin, creating polyubiquitin chains with distinct structures and functions [3] [11]. These chains can be homotypic (single linkage type), mixed, or branched (where at least one ubiquitin is modified at two different sites) [3]. UBDs decode this complex language by exhibiting a spectrum of specificity, from linkage-general binding that recognizes a common ubiquitin surface across various chain types, to highly linkage-specific interactions that discriminate between different ubiquitin chain architectures [12] [6]. This application note examines the key classes of UBDs based on their linkage recognition mechanisms and provides detailed protocols for studying their binding properties, framed within research on UBD-based affinity enrichment methods.
Key Classes of UBDs and Their Binding Mechanisms
Structural and Functional Diversity of UBD Families
UBDs encompass a wide array of structural folds, yet most target the canonical hydrophobic patch centered on Ile44 (I44 patch) on ubiquitin's β-sheet [6]. The human proteome contains more than 20 different types of UBDs, which are structurally classified into α-helical domains, zinc fingers, pleckstrin homology domains, and other folds [6]. Despite their structural diversity, these domains generally bind ubiquitin with weak affinity (typically in the 50-500 μM range for monoubiquitin), which allows for reversible and dynamic interactions necessary for cellular signaling [12].
Table 1: Major Ubiquitin-Binding Domains (UBDs) and Their Characteristics
| UBD Fold | Domain Name | Representative Protein(s) | Primary Cellular Function(s) | Typical Linkage Preference |
|---|---|---|---|---|
| α helix | UIM | Rpn10/S5a, RAP80 | Proteasomal degradation, DNA repair | General (I44 patch) |
| UBA | Rad23, Dsk2, NBR1 | Proteasome targeting, autophagy | Variable | |
| UBAN | NEMO, OPTN | NF-κB signaling | M1-linear specific | |
| Zinc finger | NZF | TAB2, TAB3, HOIL-1L, NPL4 | Kinase regulation, ERAD, MVB biogenesis | Variable (K63, M1, or general) |
| ZnF A20 | RABEX-5, A20 | Endocytosis, kinase regulation | General | |
| ZnF UBP | USP5/IsoT, HDAC6 | Proteasome function, aggresome formation | General (unanchored chains) | |
| Plekstrin Homology | PRU | RPN13 | Proteasome function | General |
| Ubc-like | UEV | Uev1/Mms2 | DNA repair, MVB biogenesis | K63-specific |
| Ubc | UBCH5C | Ubiquitin transfer | General |
Linkage-General vs. Linkage-Specific UBDs
The distinction between linkage-general and linkage-specific UBDs is fundamental to understanding ubiquitin signal interpretation:
Linkage-General UBDs: These domains primarily interact with the conserved I44 hydrophobic patch present on all ubiquitin molecules, regardless of their linkage context. They typically display low-micromolar to high-micromolar affinity for various chain types and often function as general ubiquitin sensors. Examples include many UIM (Ubiquitin-Interacting Motif) and UBA (Ubiquitin-Associated) domains [6].
Linkage-Specific UBDs: These domains contain additional structural features that enable them to recognize the unique topology of specific ubiquitin linkages. This specificity often arises from multivalent interactions where the UBD simultaneously contacts two adjacent ubiquitin moieties in a chain, with the relative orientation of these ubiquitins determining linkage preference [12]. Notable examples include:
Table 2: Quantitative Binding Affinities of Selected NZF Domains for Different Diubiquitin Linkages
| NZF Domain | K63-diUb KD (μM) | M1-diUb KD (μM) | K48-diUb KD (μM) | K11-diUb KD (μM) | Specificity Profile |
|---|---|---|---|---|---|
| TAB2 NZF | ~4 | >100 | >100 | >100 | K63-specific |
| HOIL-1L NZF | >100 | ~4 | >100 | >100 | M1-specific |
| TRABID NZF1 | >100 | >100 | >100 | ~10 (K29/K33) | K29/K33-specific |
| HOIP NZF1 | 28-48 | 28-48 | 28-48 | 28-48 | Linkage-general |
| NPL4 NZF | 113-189 | 113-189 | 113-189 | 113-189 | Linkage-general |
| ZRANB3 NZF | 28-48 | 28-48 | 28-48 | 28-48 | Linkage-general |
Experimental Protocols for Profiling UBD Specificity
Protocol 1: Comprehensive Linkage Specificity Profiling Using Surface Plasmon Resonance (SPR)
Purpose: To quantitatively determine the binding affinity and linkage preference of a UBD against all eight ubiquitin linkage types.
Materials:
- Biacore or equivalent SPR instrument
- CMS sensor chips
- Eight diubiquitin (diUb) linkage types (K6, K11, K27, K29, K33, K48, K63, M1)
- Purified UBD protein (≥95% purity)
- HBS-EP running buffer (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.005% v/v Surfactant P20, pH 7.4)
- Amine coupling reagents: NHS (N-hydroxysuccinimide), EDC (N-ethyl-N'-(3-dimethylaminopropyl)carbodiimide)
- Ethanolamine HCl
Procedure:
- Sensor Chip Preparation: Immobilize each diUb linkage type on separate flow cells of a CMS sensor chip using standard amine coupling chemistry.
- Activate the carboxymethylated dextran surface with a 7-minute injection of NHS/EDC mixture (1:1 ratio).
- Dilute diUb to 10-50 μg/mL in 10 mM sodium acetate buffer (pH 4.0-5.0) and inject until desired immobilization level is reached (typically 100-500 response units).
- Block remaining activated groups with a 7-minute injection of 1 M ethanolamine-HCl (pH 8.5).
Binding Kinetics Analysis:
- Dilute the UBD protein in HBS-EP buffer to a concentration series (typically 0.1-100 μM).
- Inject UBD concentrations over both diUb-modified and reference flow cells at a flow rate of 30 μL/min for 60-120 seconds association time, followed by 120-300 seconds dissociation time.
- Regenerate the surface with a 30-second pulse of 10 mM glycine-HCl (pH 2.0) between cycles.
Data Analysis:
- Subtract reference cell signals from diUb-modified surface responses.
- Fit resulting sensorgrams to a 1:1 Langmuir binding model to determine association (ka) and dissociation (kd) rate constants.
- Calculate equilibrium dissociation constants (KD) from the ratio kd/ka.
Interpretation: UBDs with less than 5-fold difference in KD values across linkage types are classified as linkage-general, while those showing >10-fold preference for specific linkages are considered linkage-specific [12].
Protocol 2: UBD-Based Affinity Enrichment of Ubiquitinated Substrates
Purpose: To isolate and identify ubiquitinated proteins from complex cell lysates using UBDs as affinity reagents.
Materials:
- Tandem UBD constructs (e.g., GST-tagged tandem UBA domains)
- Control non-binding mutant UBD
- Cell lysate from experimental system
- Glutathione Sepharose 4B resin
- Lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP-40, 1 mM EDTA, protease inhibitors)
- Wash buffer (same as lysis buffer with 0.1% NP-40)
- Elution buffer (50 mM Tris-HCl pH 8.0, 10 mM reduced glutathione)
- Ubiquitin linkage-specific antibodies (optional for validation)
Procedure:
- UBD Immobilization: Incubate 10-50 μg of purified tandem UBD protein with 100 μL glutathione Sepharose resin for 1 hour at 4°C. Use mutant UBD with impaired ubiquitin binding as negative control.
Sample Preparation: Prepare cell lysate from tissues or cultured cells. Clarify by centrifugation at 16,000 × g for 15 minutes. Pre-clear lysate with empty glutathione Sepharose resin for 30 minutes.
Affinity Enrichment:
- Incubate pre-cleared lysate with UBD-coupled resin for 2 hours at 4°C with gentle rotation.
- Wash resin 3-5 times with wash buffer (10 column volumes each).
- Elute bound proteins with 3-5 column volumes of elution buffer.
Downstream Analysis:
- Analyze eluates by immunoblotting with ubiquitin antibodies to confirm enrichment.
- For proteomic analysis, digest eluates with trypsin and analyze by LC-MS/MS.
- Identify ubiquitination sites by searching for GG (diGly) remnant signature (114.04 Da mass shift) on lysine residues [11].
Applications: This protocol enables identification of ubiquitinated substrates under physiological conditions without genetic manipulation, preserving native ubiquitination states [11].
Research Reagent Solutions for UBD Studies
Table 3: Essential Research Reagents for UBD-Based Affinity Enrichment
| Reagent Category | Specific Examples | Function/Application | Key Considerations |
|---|---|---|---|
| Affinity Tags | His-tag, Strep-tag, GST-tag | Purification of ubiquitinated proteins; UBD immobilization | His-tag may co-purify histidine-rich proteins; Strep-tag offers higher specificity [11] |
| Linkage-Specific DUBs | OTU family DUBs, USP53/USP54 (K63-specific) | Ubiquitin chain restriction analysis; linkage verification | USP53/54 show remarkable K63-specificity; OTUD3 prefers K48-linked chains [13] [14] |
| UBD Probes | Tandem UBA domains, linkage-specific NZFs | Affinity enrichment; ubiquitin chain detection | Tandem domains increase avidity; TAB2 NZF detects K63 chains [12] [11] |
| Activity-Based Probes | Ubiquitin-propargylamide (Ub-PA) | DUB activity profiling; active site labeling | Reacts with catalytic cysteine of active DUBs; useful for enzyme characterization [13] |
| Linkage-Specific Antibodies | K48-linkage specific, K63-linkage specific | Immunoblotting, immunofluorescence, immunoprecipitation | Enable detection of specific chain types without genetic manipulation [11] |
Schematic: Mechanisms of Linkage-Specific Ubiquitin Recognition
The diagram below illustrates how linkage-specific UBDs achieve specificity through multivalent interactions with two ubiquitin moieties simultaneously, while linkage-general UBDs primarily contact a single ubiquitin surface.
Advanced Applications: Exploiting UBD Specificity for Therapeutic Development
The precise linkage specificity of certain UBDs presents opportunities for therapeutic intervention in diseases characterized by aberrant ubiquitin signaling. For instance, the discovery that USP53 and USP54 are K63-linkage-specific deubiquitinases, with mutations in USP53 causing progressive familial intrahepatic cholestasis, highlights the potential for developing linkage-specific DUB inhibitors [13]. Furthermore, UBD-based affinity tools can be used to profile global ubiquitination changes in response to drug treatments, enabling identification of novel biomarkers and drug targets [11]. The structural insights from UBD-ubiquitin complexes, particularly the secondary interaction surfaces that confer linkage specificity, provide blueprints for designing small molecule inhibitors that disrupt pathogenic ubiquitin signaling pathways in cancer and inflammatory diseases [12] [6]. As our understanding of UBD specificity continues to evolve, particularly for branched and heterotypic ubiquitin chains, so too will opportunities for therapeutic manipulation of the ubiquitin system.
Ubiquitination is a crucial post-translational modification that regulates diverse cellular functions including protein degradation, DNA repair, cell cycle progression, and immune responses. The complexity of ubiquitin signaling—encompassing various chain topologies and linkages—presents significant analytical challenges. This application note examines ubiquitin-binding domain (UBD)-based affinity enrichment methods as essential tools for deciphering the ubiquitinome. We detail specific protocols using the high-affinity OtUBD domain, provide quantitative comparisons of enrichment methodologies, and visualize key experimental workflows. For researchers and drug development professionals, mastering these enrichment techniques is fundamental to understanding disease mechanisms and developing targeted therapies.
The ubiquitin-proteasome system represents one of the most sophisticated regulatory mechanisms in eukaryotic cells, governing protein stability, activity, localization, and interactions [15]. The term "ubiquitinome" refers to the complete set of ubiquitinated proteins within a biological system at a specific timepoint. This dynamic landscape provides critical insights into cellular status, particularly under pathological conditions.
Ubiquitination involves the covalent attachment of ubiquitin—a 76-amino acid protein—to substrate proteins via a sequential enzymatic cascade involving E1 activating, E2 conjugating, and E3 ligase enzymes [11] [15]. The complexity arises from ubiquitin's own modification potential: it contains seven lysine residues (K6, K11, K27, K29, K33, K48, K63) and an N-terminal methionine (M1) that can form various polyubiquitin chains with distinct biological functions [15]. For instance, K48-linked chains primarily target substrates for proteasomal degradation, while K63-linked chains often function in signaling pathways such as DNA repair and inflammation [11]. The specificity of these signals is decoded by ubiquitin-binding domains (UBDs), protein modules that recognize and bind non-covalently to ubiquitin [7].
Dysregulation of ubiquitination pathways is implicated in numerous diseases. In cancer, altered E3 ligase or deubiquitinase (DUB) activity can lead to uncontrolled proliferation or evasion of cell death [15]. Neurodegenerative disorders like Alzheimer's disease feature abnormal accumulation of ubiquitinated proteins, exemplified by K48-linked polyubiquitination of tau proteins [11]. These connections make comprehensive ubiquitinome analysis not merely an academic exercise but a critical requirement for understanding disease pathogenesis and identifying therapeutic targets.
Methodological Approaches for Ubiquitinome Enrichment
Several strategies have been developed to enrich ubiquitinated proteins or peptides from complex biological samples, each with distinct advantages and limitations. The following table summarizes the primary methodologies:
Table 1: Comparison of Ubiquitinome Enrichment Methods
| Method | Principle | Advantages | Limitations | Typical Applications |
|---|---|---|---|---|
| Ubiquitin Tagging | Expression of epitope-tagged (His, HA, Flag, Strep) ubiquitin in cells [11] [16] | Relatively low-cost; technically accessible; good for cell culture systems | May alter ubiquitin structure/function; cannot be used in human tissues; potential spurious ubiquitination patterns [11] [16] | Screening ubiquitinated substrates in engineered cell lines; initial discovery studies |
| Antibody-based Enrichment | Immunoaffinity purification using ubiquitin-specific antibodies (e.g., P4D1, FK1/FK2) or linkage-specific antibodies [11] [16] | Works with endogenous ubiquitin; applicable to clinical samples and animal tissues; linkage-specific versions available [11] | High cost; potential non-specific binding; may have preference for certain ubiquitin conformations [11] [16] | Disease mechanism studies using patient samples; linkage-specific ubiquitination analysis |
| UBD-based Affinity | Utilization of ubiquitin-binding domains (e.g., OtUBD, TUBEs) as affinity reagents [17] [11] | High affinity for endogenous ubiquitin; captures both mono- and polyubiquitinated proteins; versatile application across sample types [17] | Requires protein engineering; optimization needed for different biological contexts; potential co-purification of interacting proteins under native conditions [17] [11] | Comprehensive ubiquitinome profiling; distinction between covalent ubiquitination and non-covalent interactions |
| diGLY Remnant Peptide | Immunoaffinity purification of tryptic peptides containing Gly-Gly remnant on modified lysines after ubiquitin cleavage [16] [18] | High specificity; enables precise ubiquitination site mapping; compatible with quantitative proteomics | Loss of information about ubiquitin chain topology; cannot distinguish ubiquitin from NEDD8/ISG15 modifications; requires specialized antibodies [16] | High-throughput site-specific ubiquitination analysis; quantitative studies of ubiquitination dynamics |
Among these approaches, UBD-based methods offer particular advantages for comprehensive ubiquitinome profiling. Traditional single UBDs often suffer from low affinity, with dissociation constants typically in the low to mid μM range [7]. This limitation has been addressed through engineered solutions such as tandem-repeated UBDs (TUBEs) and the recently characterized OtUBD—a high-affinity ubiquitin-binding domain derived from Orientia tsutsugamushi with dissociation constants in the low nanomolar range [17]. The exceptional binding affinity of OtUBD enables efficient capture of diverse ubiquitin conjugates, including both mono- and polyubiquitinated proteins that constitute a large fraction of the ubiquitinome in mammalian cells [17].
Protocol: OtUBD-Based Enrichment of Ubiquitinated Proteins
This section provides a detailed methodology for using the OtUBD domain to enrich ubiquitinated proteins from biological samples, adapted from established protocols [17].
Reagent Preparation
- Lysis Buffers: Prepare both native (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM DTT, 10 mM N-ethylmaleimide [NEM], protease inhibitors) and denaturing (6 M guanidine-HCl, 100 mM NaH₂PO₄, 10 mM Tris–HCl pH 8.0, 10 mM NEM, 5 mM imidazole) formulations
- OtUBD Affinity Resin: Couple recombinant Cys-His₆-OtUBD to SulfoLink coupling resin according to manufacturer's instructions
- Wash Buffers: Prepare low-stringency (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.1% Triton X-100) and high-stringency (50 mM Tris–HCl pH 7.5, 500 mM NaCl, 0.5% Triton X-100) buffers
- Elution Buffer: 50 mM Tris–HCl pH 7.5, 150 mM NaCl, 2% SDS, 10 mM DTT
Step-by-Step Procedure
Sample Preparation:
- For yeast cells: Resuspend cell pellet in lysis buffer and disrupt using acid-washed glass beads with vortexing
- For mammalian cells: Directly lyse cells in appropriate buffer using a syringe and needle
- Clarify lysates by centrifugation at 20,000 × g for 15 minutes at 4°C
- Determine protein concentration using Bradford or BCA assay
Affinity Enrichment:
- Incubate clarified lysate (typically 1-5 mg total protein) with OtUBD affinity resin (50 μL bed volume per mg protein) for 2 hours at 4°C with end-over-end mixing
- Pellet resin by gentle centrifugation (500 × g for 5 minutes) and carefully remove supernatant
Washing:
- Wash resin sequentially with:
- 10 bed volumes of low-stringency wash buffer
- 10 bed volumes of high-stringency wash buffer
- 10 bed volumes of 50 mM Tris–HCl pH 7.5
- For denaturing conditions: Include additional washes with 6 M urea in 50 mM Tris–HCl pH 7.5
- Wash resin sequentially with:
Elution:
- Elute bound proteins with 2-3 bed volumes of elution buffer at 95°C for 10 minutes
- Collect eluate and repeat elution once
- Combine eluates for downstream analysis
Downstream Applications:
- For immunoblotting: Separate proteins by SDS-PAGE and transfer to PVDF membrane
- For proteomics: Process samples using filter-aided sample preparation (FASP) or similar methods prior to LC-MS/MS analysis
Critical Protocol Notes
- Native vs. Denaturing Conditions: The native workflow enriches both directly ubiquitinated proteins and proteins that interact with ubiquitin or ubiquitinated proteins, while the denaturing workflow specifically isolates covalently ubiquitinated proteins [17]
- Protease Inhibition: Include N-ethylmaleimide (NEM) or iodoacetamide in all buffers to inhibit deubiquitinases and preserve ubiquitin conjugates
- Quality Control: Validate enrichment efficiency by immunoblotting with anti-ubiquitin antibodies (e.g., P4D1, E412J)
- Scale Considerations: For proteomic applications, scale up protein input accordingly (typically 5-10 mg for deep ubiquitinome coverage)
The following diagram illustrates the key decision points in the OtUBD enrichment workflow:
Research Reagent Solutions
Successful implementation of UBD-based enrichment protocols requires specific reagents and tools. The following table details essential research solutions for ubiquitinome studies:
Table 2: Essential Research Reagents for UBD-Based Ubiquitinome Studies
| Reagent/Category | Specific Examples | Function and Application | Considerations for Use |
|---|---|---|---|
| UBD Affinity Reagents | OtUBD resin [17], Tandem UBDs (TUBEs) [11] | High-affinity capture of ubiquitinated proteins from complex lysates | OtUBD offers nanomolar affinity and recognizes both mono- and polyubiquitin; TUBEs preferentially bind polyubiquitin chains |
| Ubiquitin Antibodies | P4D1 [17], FK1/FK2 [11], E412J [17], linkage-specific antibodies [11] [16] | Detection and validation of ubiquitinated proteins; some can be used for enrichment | Linkage-specific antibodies enable study of chain topology; validation required for each application |
| Protease Inhibitors | N-ethylmaleimide (NEM) [17], phenylmethylsulfonyl fluoride (PMSF) [17], complete EDTA-free protease inhibitor cocktail [17] | Preserve ubiquitin conjugates by inhibiting deubiquitinases and proteases | NEM is essential for DUB inhibition; include in all lysis and binding buffers |
| Mass Spectrometry Reagents | diGLY remnant antibodies [16] [18], SDC lysis buffer [18], chloroacetamide (CAA) [18] | Ubiquitination site mapping via proteomics | SDC lysis with CAA increases ubiquitin site coverage compared to urea buffers [18] |
| Expression Plasmids | pRT498-OtUBD [17], pET21a-cys-His6-OtUBD [17] | Recombinant production of UBDs for resin generation | Available through Addgene for protocol implementation |
Ubiquitinome Analysis by Mass Spectrometry
Mass spectrometry has revolutionized ubiquitinome studies by enabling systematic identification and quantification of ubiquitination sites. The diGLY remnant profiling approach leverages the fact that tryptic digestion of ubiquitinated proteins leaves a characteristic Gly-Gly modification on the modified lysine residue [16]. Recent methodological advances have significantly improved the depth and precision of ubiquitinome coverage:
Table 3: Quantitative Comparison of Ubiquitinome Profiling by Mass Spectrometry
| Methodological Parameter | Traditional DDA with Urea Lysis | Improved DDA with SDC Lysis | DIA-MS with SDC Lysis |
|---|---|---|---|
| Typical K-GG Peptide Identifications | ~19,400 peptides [18] | ~26,750 peptides [18] | ~68,400 peptides [18] |
| Reproducibility (CV < 20%) | Lower [18] | Moderate improvement [18] | High (median CV ~10%) [18] |
| Protein Input Requirements | High (often >5 mg) [16] | Moderate (2 mg) [18] | Flexible (0.5-4 mg tested) [18] |
| Quantitative Precision | Limited by missing values [18] | Improved but still significant missing values [18] | Excellent (>68,000 peptides quantifiable across replicates) [18] |
| Technical Implementation | Established workflows | Requires protocol adaptation | Requires specialized DIA methods and analysis software (DIA-NN) [18] |
The implementation of data-independent acquisition (DIA) mass spectrometry, coupled with improved sample preparation using sodium deoxycholate (SDC)-based lysis and chloroacetamide (CAA) for rapid cysteine alkylation, has dramatically enhanced ubiquitinome profiling [18]. This optimized workflow more than triples identification numbers compared to traditional data-dependent acquisition (DDA) approaches while significantly improving quantitative precision [18].
The following diagram illustrates the integrated workflow combining OtUBD enrichment with advanced mass spectrometry for comprehensive ubiquitinome analysis:
Applications in Disease Research and Therapeutic Development
The strategic importance of ubiquitinome enrichment becomes evident when examining its applications in disease mechanism studies and drug development. Several key areas exemplify this translational potential:
Target Deconvolution for DUB Inhibitors
Comprehensive ubiquitinome profiling enables rapid mode-of-action studies for drugs targeting deubiquitinases (DUBs) or ubiquitin ligases. When applied to USP7 inhibition, time-resolved ubiquitinome analysis revealed that while ubiquitination of hundreds of proteins increased within minutes of inhibitor treatment, only a small subset of these targets underwent degradation [18]. This distinction between regulatory and degradative ubiquitination provides critical insights for drug development, suggesting that monitoring both ubiquitination changes and corresponding protein abundance is essential for complete pharmacological assessment.
Biomarker Discovery in Neurodegenerative Diseases
Ubiquitinome analysis of patient-derived samples has identified disease-specific ubiquitination signatures. In Alzheimer's disease research, specialized antibodies against K48-linked polyubiquitin chains revealed abnormal accumulation of K48-polyubiquitinated tau proteins [11]. Such linkage-specific ubiquitination patterns may serve as diagnostic biomarkers or therapeutic response indicators, highlighting the value of enrichment methods that preserve chain topology information.
Personalized Medicine Approaches
The ability to profile ubiquitination changes in response to targeted therapies creates opportunities for treatment stratification. As many oncogenic signaling pathways are regulated through ubiquitination, monitoring dynamic changes in the ubiquitinome following drug treatment could identify predictive biomarkers of response and resistance mechanisms [15]. UBD-based enrichment methods applied to clinical specimens could therefore guide therapeutic decisions in precision oncology.
Ubiquitinome enrichment represents more than a technical procedure—it is a biological imperative for advancing our understanding of disease mechanisms and developing targeted therapies. UBD-based affinity methods, particularly those utilizing high-affinity domains like OtUBD, provide powerful tools for comprehensive ubiquitinome characterization. When integrated with advanced mass spectrometry techniques such as DIA, these approaches enable unprecedented depth and precision in monitoring ubiquitination dynamics. For researchers and drug development professionals, mastering these methodologies is essential for deciphering the complex language of ubiquitin signaling in health and disease.
The UBD Toolbox: Protocols and Applications for Ubiquitin Enrichment
The ubiquitin-proteasome system (UPS) represents a crucial regulatory pathway in eukaryotic cells, controlling protein stability, activity, and localization through the covalent attachment of ubiquitin [19]. This post-translational modification generates diverse ubiquitin conjugates, including monoubiquitination, multiple monoubiquitination, and polyubiquitin chains with various linkage types, each encoding distinct cellular signals [20]. The complexity of ubiquitin signaling presents significant challenges for its study, necessitating robust methods for the specific isolation and detection of ubiquitinated proteins from complex biological samples [19].
Among the methodologies developed for ubiquitin research, affinity enrichment techniques using ubiquitin-binding domains (UBDs) have emerged as powerful tools [19]. These domains, which naturally occur in many proteins involved in ubiquitin signaling, recognize and bind to ubiquitin modifications with varying specificities and affinities [20]. Tandem-repeated Ubiquitin-Binding Entities (TUBEs) represent an engineered advancement in this field, harnessing the strength of multiple UBDs to overcome limitations of traditional approaches such as immunoprecipitation with ubiquitin antibodies or epitope-tagged ubiquitin [21].
Principles of TUBE Technology
Fundamental Design and Mechanism
Tandem Ubiquitin Binding Entities are engineered protein domains that incorporate multiple ubiquitin-binding domains (UBDs) within a single polypeptide chain [22]. This design strategically addresses the fundamental challenge of low-affinity binding associated with individual UBDs by creating an avidity effect, where the simultaneous interaction of multiple UBDs with a polyubiquitin chain results in dramatically enhanced binding strength [21] [19]. LifeSensors, a pioneer in TUBE technology, has developed TUBEs that bind to polyubiquitin chains with dissociation constants (Kd) in the nanomolar range, typically between 1-10 nM [21].
The molecular architecture of TUBEs enables them to recognize the characteristic structural features of ubiquitin chains. Most UBDs interact with the hydrophobic patch on ubiquitin formed by residues Leu8, Ile44, and Val70 [20]. By positioning multiple UBDs in tandem, TUBEs achieve high-affinity binding that far exceeds that of natural UBD-containing proteins or conventional ubiquitin antibodies. This design principle effectively circumvents the need for immunoprecipitation of overexpressed epitope-tagged ubiquitin or the use of ubiquitin antibodies, which are notoriously non-selective and can lead to artifacts [21].
Key Functional Properties
Beyond their exceptional binding affinity, TUBEs possess several remarkable functional properties that make them invaluable for ubiquitin research. Most notably, TUBEs have been demonstrated to protect ubiquitylated proteins from both deubiquitylation and proteasome-mediated degradation, even in the absence of the deubiquitinase (DUB) and proteasome inhibitors that are normally required to preserve ubiquitin signals in cell lysates [21]. This protective function significantly enhances the recovery of labile ubiquitin conjugates that might otherwise be lost during sample preparation.
Additionally, TUBEs exhibit versatile recognition capabilities for different ubiquitin chain architectures. While individual UBDs often show preferences for specific chain types, the tandem arrangement in TUBEs can be engineered to create either broad-specificity or chain-selective reagents. This flexibility allows researchers to either capture the global ubiquitinome or focus on specific ubiquitin signaling pathways [21] [22].
Table 1: Key Properties and Advantages of TUBE Technology
| Property | Technical Advantage | Application Benefit |
|---|---|---|
| Nanomolar affinity (Kd = 1-10 nM) | Strong polyubiquitin chain binding | Enhanced sensitivity for low-abundance ubiquitinated proteins |
| Protection from DUBs/proteasomes | Stabilizes ubiquitin conjugates without inhibitors | Preserves labile ubiquitination events; simplifies experimental procedures |
| Chain-type selectivity | Can be engineered for specific linkages or pan-specific recognition | Enables study of specific ubiquitin signaling pathways or global ubiquitination |
| Adaptable detection modalities | Compatible with various tags (e.g., TAMRA, biotin) | Facilitates diverse applications: pulldowns, Western blotting, imaging, HTS |
Comparative Analysis of Ubiquitin Enrichment Methods
To fully appreciate the advantages of TUBE technology, it is essential to contextualize it within the broader landscape of ubiquitin enrichment methodologies. Currently, three primary approaches dominate the field: ubiquitin antibodies, tagged ubiquitin systems, and UBD-based methods including TUBEs [19].
Ubiquitin antibody-based enrichment utilizes antibodies such as P4D1 or FK1/FK2 that recognize all ubiquitin linkages, or linkage-specific antibodies targeting M1, K11, K27, K48, or K63 chains [19] [23]. While these antibodies enable the study of endogenous ubiquitination without genetic manipulation, they suffer from high cost, potential non-specific binding, and variable specificity depending on the supplier [21] [19].
Tagged ubiquitin approaches (e.g., His-, FLAG-, or Strep-tagged ubiquitin) involve expressing tagged ubiquitin in cells, allowing affinity purification of ubiquitinated proteins using corresponding resin systems [19]. Although widely used, these methods introduce artificial genetic constructs that may not fully recapitulate endogenous ubiquitin biology and cannot be applied to clinical tissue samples [19].
UBD-based methods represent a more recent development, leveraging natural ubiquitin-recognition domains. Single UBDs initially showed promise but were limited by low affinity. TUBEs address this limitation through their tandem domain architecture [19]. Another UBD-based tool recently described is OtUBD, a high-affinity ubiquitin-binding domain derived from Orientia tsutsugamushi that can enrich both mono- and poly-ubiquitinated proteins, contrasting with TUBEs' preference for polyubiquitin chains [4] [24].
Table 2: Comparison of Ubiquitinated Protein Enrichment Methods
| Method | Sensitivity | Specificity | Endogenous Application | Key Limitations |
|---|---|---|---|---|
| Ubiquitin Antibodies | Moderate | Variable; linkage-specific options available | Yes | High cost; potential non-specific binding; lot-to-lot variability |
| Tagged Ubiquitin | High | High for tagged ubiquitin | No (requires genetic manipulation) | Cannot use on tissues; may alter native ubiquitin function |
| Single UBDs | Low | Variable | Yes | Low affinity limits utility for comprehensive ubiquitinome studies |
| TUBEs | High (nanomolar Kd) | High; pan-selective or chain-specific options | Yes | Lower efficiency for monoubiquitinated proteins |
| OtUBD | High (nanomolar Kd) | High for both mono- and polyubiquitin | Yes | Recently developed; less established protocol |
TUBE Reagent Systems and Specifications
TUBE technology has evolved to include a diverse repertoire of reagents tailored for different experimental needs. These can be broadly categorized into pan-selective TUBEs that recognize all ubiquitin chain types, and chain-selective TUBEs that target specific linkages [21] [22].
Pan-TUBEs exhibit broad specificity for polyubiquitin chains regardless of linkage type, making them ideal for global ubiquitinome profiling and proteomic studies aimed at discovering novel ubiquitination events. These reagents typically incorporate UBDs with general ubiquitin-binding properties, such as domains from proteins like Rabex-5 [21].
Chain-selective TUBEs have been engineered to recognize specific ubiquitin chain linkages, enabling researchers to focus on particular ubiquitin-dependent pathways. LifeSensors has developed several chain-selective TUBEs, including K48-specific TUBEs (marketed as K48 HF TUBEs) that target the canonical degradation signal, K63-specific TUBEs for studying DNA repair, endocytosis, and NF-κB signaling pathways, and M1-specific TUBEs for investigating linear ubiquitination in inflammatory signaling [21].
These TUBE reagents are available in various formats to support diverse applications. Tagged TUBEs (e.g., His-tagged, GST-tagged, or TAMRA-labeled) facilitate different detection and purification strategies. For instance, TAMRA-TUBE2 features a fluorophore attached to the fusion tag without interfering with ubiquitin binding, enabling imaging applications to study intracellular ubiquitination dynamics [21]. Immobilized TUBEs are conjugated to solid supports such as agarose beads, streamlining pull-down experiments for proteomic analyses [21].
Table 3: Research Reagent Solutions for TUBE-Based Experiments
| Reagent | Composition/Format | Primary Function | Example Applications |
|---|---|---|---|
| Pan-TUBEs | Tandem UBDs with broad specificity | Global capture of polyubiquitinated proteins | Ubiquitinome profiling by mass spectrometry; protection assays |
| Chain-Selective TUBEs | Engineered UBDs with linkage preference | Isolation of specific ubiquitin chain types | Studying K48-linked degradation or K63-linked signaling pathways |
| TAMRA-TUBE2 | TUBE with fluorophore on fusion tag (Ex. 540 nm/Em. 578 nm) | Visualization of ubiquitin conjugates | Imaging ubiquitination dynamics in cells; fluorescent detection assays |
| Immobilized TUBEs | TUBEs conjugated to agarose beads | Affinity capture of ubiquitinated proteins | Pull-down experiments; sample preparation for proteomics |
| OtUBD Resin | High-affinity UBD from O. tsutsugamushi on resin | Enrichment of mono- and polyubiquitinated proteins | Proteomics under native or denaturing conditions; ubiquitin interactome studies |
Detailed Experimental Protocols
TUBE-Based Pull-Down for Ubiquitinated Protein Enrichment
The following protocol describes a standardized procedure for enriching ubiquitinated proteins from cell lysates using TUBE technology, particularly applicable for downstream applications such as mass spectrometry proteomics, Western blotting, or imaging [21].
Reagents and Materials
- Cell lysate: Prepare using appropriate lysis buffer (e.g., RIPA buffer) supplemented with protease inhibitors. Although TUBEs offer protection, including 1-10 mM N-ethylmaleimide (NEM) is recommended to inhibit deubiquitinases [21] [24].
- TUBE reagent: Select appropriate TUBE based on experimental needs (pan-selective or chain-selective; His-tagged for nickel resin or agarose-immobilized) [21].
- Binding buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5-1% NP-40 or Triton X-100, with optional 1-10 mM NEM [24].
- Elution buffer: 1X SDS-PAGE loading buffer with 100 mM DTT for direct Western analysis, or competitive elution buffers with free ubiquitin for native elution [24].
Step-by-Step Procedure
- Lysate Preparation: Harvest cells and lyse in appropriate buffer. Clear lysate by centrifugation at 14,000 × g for 15 minutes at 4°C. Determine protein concentration using Bradford or BCA assay [24].
- TUBE Incubation: Incubate 500 μg - 1 mg of total cell lysate with 2-5 μg of appropriate TUBE reagent for 2-4 hours at 4°C with gentle rotation [21].
- Capture: For tagged TUBEs, add appropriate affinity resin (e.g., Ni-NTA agarose for His-tagged TUBEs) and incubate for 1-2 hours at 4°C with rotation [24].
- Washing: Pellet resin and wash 3-4 times with binding buffer. Increase stringency with higher salt (up to 300 mM NaCl) in final wash if needed to reduce non-specific binding [24].
- Elution: Elute bound proteins by adding 1X SDS-PAGE loading buffer with 100 mM DTT and heating at 95°C for 5-10 minutes, or use competitive elution with free ubiquitin (0.1-1 mg/mL) for native applications [24].
- Analysis: Proceed with Western blotting using appropriate ubiquitin antibodies (e.g., P4D1, FK2, or E412J) or process for mass spectrometry analysis [24].
Denatured-Refolded Ubiquitinated Sample Preparation (DRUSP) with Tandem Hybrid UBD
The DRUSP protocol represents an advanced methodology that combines strong denaturation with refolding to significantly enhance the efficiency and reproducibility of ubiquitinomics research [5].
- Denaturing Extraction: Lyse cells or tissues using strongly denaturing buffers (e.g., high urea or SDS concentrations) to efficiently extract ubiquitinated proteins while completely inactivating DUBs and proteasomes [5].
- Filter-Based Refolding: Dilute denatured lysates and use filter-based devices to gradually remove denaturants, allowing ubiquitinated proteins to refold into native conformations recognizable by UBDs [5].
- TUBE Enrichment: Apply refolded samples to TUBE affinity resin for specific capture of ubiquitinated proteins [5].
- Proteomic Analysis: Process enriched ubiquitinated proteins for LC-MS/MS analysis to identify ubiquitination sites and quantify changes [5].
Key Advantages
- Enhanced signal: DRUSP yields significantly stronger ubiquitin signals, nearly three times greater than control methods under native conditions [5].
- Improved reproducibility: The denaturing extraction minimizes variability caused by differential DUB and proteasome activities across samples [5].
- Comprehensive coverage: This method enables extraction of ubiquitinated proteins that might be insoluble or poorly extracted under native conditions [5].
- Versatility: DRUSP works effectively with both pan-selective and chain-specific UBDs, allowing linkage-specific ubiquitinome profiling [5].
TUBE-Based Western Blot Detection
TUBEs can serve as alternative detection reagents to ubiquitin antibodies in Western blotting, offering enhanced sensitivity and specificity for polyubiquitinated proteins [22].
- Electrophoresis and Transfer: Separate proteins by SDS-PAGE and transfer to PVDF membrane using standard protocols [24].
- Blocking: Block membrane with 5% non-fat milk or BSA in TBST for 1 hour at room temperature [24].
- TUBE Probing: Incubate membrane with appropriate tagged TUBE (1-2 μg/mL in blocking buffer) for 2 hours at room temperature or overnight at 4°C [21] [22].
- Tag Detection: For His-tagged TUBEs, incubate with anti-His antibody (1:2000-1:5000 dilution) for 1 hour. For TAMRA-TUBE, direct fluorescence detection can be used [21].
- Secondary Detection: Incubate with HRP-conjugated secondary antibody if necessary and develop with ECL reagent [24].
Applications in Drug Discovery and Targeted Protein Degradation
TUBE technology has found particularly valuable applications in the rapidly expanding field of targeted protein degradation (TPD), facilitating the development of novel therapeutic modalities such as PROTACs (PROteolysis TArgeting Chimeras) and molecular glues [21] [22].
Monitoring PROTAC Efficiency
PROTAC molecules function by inducing proximity between an E3 ubiquitin ligase and a target protein of interest, leading to target ubiquitination and subsequent proteasomal degradation [22]. TUBEs provide a direct means to monitor the efficiency of this process through multiple applications:
High-Throughput Screening Platforms: LifeSensors has developed TUBE-based assays configured in microtiter plate formats to rapidly quantify polyubiquitination of target proteins in response to PROTAC treatment [21]. These platforms enable rank-order potency assessment of candidate molecules and facilitate structure-activity relationship studies throughout the drug discovery pipeline [21].
Mechanistic Studies: TUBEs enable researchers to distinguish between different mechanisms of action in TPD. For instance, K48-specific TUBEs can confirm the formation of degradative ubiquitin chains, while protection from deubiquitination allows stabilization of transient ubiquitination events that might be difficult to capture otherwise [21] [22].
Molecular Glue Characterization
Molecular glues perform similar functions to PROTACs by inducing neomorphic interactions between E3 ligases and target proteins but typically have lower molecular weights and are discovered through traditional screening approaches [21]. TUBEs provide valuable tools for characterizing molecular glue mechanisms by:
- Ubiquitination confirmation: Direct detection of target protein ubiquitination following molecular glue treatment [21].
- Temporal monitoring: Tracking the kinetics of ubiquitination and subsequent degradation in cellular models [21].
- Linkage determination: Using chain-selective TUBEs to identify the types of ubiquitin chains assembled on target proteins [21].
The following diagram illustrates the application of TUBE technology in monitoring PROTAC-induced ubiquitination:
Emerging Methodologies and Future Perspectives
While TUBE technology represents a significant advancement in ubiquitin research, continuous innovation in the field has yielded complementary approaches that address specific limitations. The recent development of OtUBD exemplifies this progress, offering a high-affinity ubiquitin-binding domain derived from Orientia tsutsugamushi that demonstrates exceptional binding properties [4] [25].
OtUBD Technology
The OtUBD domain exhibits several distinctive characteristics that complement TUBE technology:
Exceptional Affinity: OtUBD binds monoubiquitin with an unprecedented dissociation constant of approximately 5 nM, making it one of the highest affinity UBDs identified to date [25].
Broad Substrate Recognition: Unlike TUBEs that preferentially recognize polyubiquitin chains, OtUBD efficiently enriches both mono- and poly-ubiquitinated proteins from complex biological samples [4] [24].
Structural Plasticity: Biophysical studies reveal that OtUBD undergoes a pronounced structural transition upon ubiquitin binding, transitioning from a poorly folded to well-ordered state, which may contribute to its exceptional binding properties [25].
Versatile Applications: OtUBD has been successfully utilized in various experimental workflows, including immunoblotting, differential proteomics, and UbiCREST (ubiquitin chain restriction) analyses [4].
Protocol for OtUBD-Mediated Enrichment
The following protocol outlines the key steps for utilizing OtUBD affinity resin to enrich ubiquitinated proteins from yeast or mammalian cell lysates [4] [24]:
- Resin Preparation: Express and purify recombinant OtUBD using pET21a-cys-His6-OtUBD plasmid (Addgene #190091). Couple to SulfoLink coupling resin according to manufacturer's instructions [24].
- Lysate Preparation: Prepare cell lysates under either native (for ubiquitin interactome) or denaturing conditions (for covalent ubiquitinome) using appropriate buffer formulations [4].
- Enrichment: Incubate cleared lysates with OtUBD resin for 2-4 hours at 4°C. Include controls with free ubiquitin for competition experiments to verify specificity [24].
- Washing and Elution: Wash resin with appropriate buffers (varying stringency based on native vs. denaturing conditions) and elute bound proteins with SDS-PAGE sample buffer or competitive elution with free ubiquitin [24].
- Downstream Analysis: Process eluates for Western blotting with anti-ubiquitin antibodies or for LC-MS/MS analysis to identify ubiquitination sites [4] [24].
Future Directions
The continued evolution of UBD-based affinity tools promises to further advance ubiquitin research. Key areas of development include:
Enhanced Specificity: Engineering UBDs with improved linkage specificity for less common ubiquitin chain types (e.g., K6, K27, K29, K33) to facilitate study of their biological functions [19].
Multiplexed Analysis: Developing UBD panels that enable parallel analysis of multiple ubiquitin chain types from limited sample quantities, particularly relevant for clinical specimens [5].
Integration with Single-Cell Technologies: Adapting UBD-based enrichment for compatibility with emerging single-cell proteomic platforms to explore ubiquitination heterogeneity in complex tissues [19].
Chemical Biology Tools: Creating bifunctional UBD compounds that can crosslink to ubiquitinated proteins or associated complexes for structural studies and interaction proteomics [22].
In conclusion, TUBE technology and related UBD-based affinity methods have revolutionized the study of protein ubiquitination by providing robust, specific, and versatile tools for enrichment and detection. As these technologies continue to evolve alongside advances in mass spectrometry and chemical biology, they will undoubtedly yield deeper insights into the complex landscape of ubiquitin signaling in health and disease.
The ubiquitin code represents one of the most complex post-translational regulatory systems in eukaryotic cells, where diverse ubiquitin chain architectures—differing in linkage type, length, and branching—encode precise biological outcomes for modified substrates [26] [27]. Ubiquitin-binding domains (UBDs) serve as fundamental decoders of this complex language, enabling cellular machinery to interpret ubiquitin signals and execute appropriate downstream functions. Among the eight possible ubiquitin linkage types, lysine 48-linked (K48) and lysine 63-linked (K63) polyubiquitin chains constitute the most abundant and well-characterized signals, with K48 chains primarily targeting substrates for proteasomal degradation and K63 chains regulating non-proteolytic processes including DNA repair, signaling, and trafficking [26] [28]. The remaining "atypical" linkages (K6, K11, K27, K29, K33) and methionine 1-linked (M1) linear chains add further layers of complexity to this signaling system, creating an extensive vocabulary of biological responses [28] [27].
Linkage-specific UBDs have emerged as indispensable tools for ubiquitin research, enabling selective enrichment and analysis of distinct ubiquitin chain types from complex biological samples. Traditional methods for studying protein ubiquitination, including epitope-tagged ubiquitin expression and anti-ubiquitin antibodies, present significant limitations in distinguishing between chain architectures and often fail to detect monoubiquitination or non-canonical modifications [17] [29]. The development of UBD-based affinity reagents with defined linkage preferences has revolutionized our ability to decipher the ubiquitin code by providing researchers with targeted approaches to isolate and characterize specific ubiquitin signals. This application note details contemporary methodologies and tools for linkage-specific ubiquitin enrichment, with particular emphasis on K48, K63, and atypical chain recognition, providing researchers with practical protocols for implementing these techniques in their experimental systems.
The Ubiquitin Code: Complexity and Biological Significance
Ubiquitin modifications exhibit remarkable structural diversity, functioning as a sophisticated molecular language that directs cellular processes. As illustrated below, this complexity encompasses multiple dimensions of variability, from single ubiquitin modifications to complex branched chains.
Figure 1: The Complexity of Ubiquitin Modifications. Ubiquitin signals range from single modifications to complex chains with distinct biological functions. Linkage types are color-coded: red (K48), green (K63), blue (atypical), and dark gray (M1-linear).
The functional consequences of ubiquitination extend far beyond the canonical role of K48-linked chains in proteasomal degradation. K63-linked chains serve critical roles in inflammatory signaling, DNA damage repair, and endocytic trafficking [26] [28]. Atypical chains have more specialized functions: K6-linked chains regulate mitophagy and the DNA damage response; K11-linked chains control cell cycle progression; K27- and K29-linked chains function in immune signaling and kinase regulation; while K33-linked chains influence trafficking processes [28]. Branched ubiquitin chains, in which a single ubiquitin moiety is modified at multiple sites, represent an additional layer of complexity, with K48/K63-branched chains comprising approximately 20% of all K63 linkages in cells and potentially serving as enhanced degradation signals or regulatory switches [26] [3]. This architectural diversity enables exquisite specificity in cellular regulation but presents significant challenges for experimental dissection, necessitating sophisticated tools for chain-type-specific isolation and analysis.
Current Landscape of Linkage-Specific Ubiquitin Binding Domains
The recognition of specific ubiquitin chain architectures is mediated by UBDs with precise structural preferences. Recent advances have identified and characterized numerous linkage-specific UBDs, while also revealing that some compact UBDs achieve specificity through multivalent interactions with both ubiquitin and the modified substrate [8]. The table below summarizes key linkage-specific UBDs and their functional characteristics.
Table 1: Linkage-Specific Ubiquitin-Binding Domains (UBDs) and Their Characteristics
| UBD/Reagent | Linkage Specificity | Affinity/KD | Key Features | Applications |
|---|---|---|---|---|
| OtUBD | Pan-ubiquitin (mono & poly) | Low nM range [17] | High affinity, recognizes monoubiquitination & non-canonical sites [29] | Ubiquitinome profiling, interactome analysis [17] |
| NZF Domains | Variable (K63, K6, or broad) | Weak (μM) individually [8] | Compact (~30 aa), secondary interfaces, multivalent binding [8] | Chain linkage analysis, targeted enrichment |
| UBD in USP54 | K63-specific | N/A | Cryptic S2 ubiquitin-binding site [13] | K63-chain cleavage, linkage-specific DUB assays |
| TAB2 NZF | K63 & phosphorylated Ser65-K6 | N/A | Recognizes ubiquitin phosphorylation status [8] | Mitophagy studies, phospho-ubiquitin detection |
| HOIP NZF1 | Subsite-dependent (ubiquitinated NEMO/optineurin) | N/A | Binds ubiquitinated substrates specifically [8] | Linear ubiquitination analysis, LUBAC signaling |
| UBD in USP53 | K63-specific | N/A | K63-specific S2 site, en bloc deubiquitination [13] | K63-chain editing, linkage-specific DUB assays |
The high-affinity OtUBD derived from Orientia tsutsugamushi represents a significant advancement in ubiquitin enrichment technology [17] [29]. Unlike tandem ubiquitin-binding entities (TUBEs) that primarily recognize polyubiquitin chains with low efficiency for monoubiquitination, OtUBD exhibits nanomolar affinity for both mono- and polyubiquitinated proteins, enabling comprehensive ubiquitinome profiling [29]. Furthermore, OtUBD can detect non-canonical ubiquitination sites (serine, threonine, cysteine, and N-terminal residues) that are often missed by diGly remnant antibodies [29]. This versatility makes OtUBD particularly valuable for discovering novel ubiquitination types and profiling complete ubiquitinomes across experimental conditions.
NZF domains exemplify how small UBDs achieve functional diversity despite their compact size. While some NZF domains exhibit clear linkage preferences—such as the TAB2 NZF domain that recognizes both K63 linkages and K6-linked chains phosphorylated at Ser65—many NZF domains display surprisingly broad linkage recognition [8]. Recent research indicates that these domains may achieve specificity through secondary interaction interfaces that simultaneously contact both ubiquitin and the modified substrate itself [8]. This mechanism is exemplified by the NZF1 domain of HOIP, which preferentially binds site-specifically ubiquitinated forms of NEMO and optineurin rather than unanchored ubiquitin chains [8].
The recent discovery that USP53 and USP54, previously annotated as catalytically inactive pseudodeubiquitinases, are actually K63-specific deubiquitinases with cryptic S2 ubiquitin-binding sites further expands the toolkit for K63-chain research [13]. These enzymes exhibit remarkable linkage specificity, with USP54 cleaving within K63-linked chains while USP53 performs K63-linkage-directed en bloc deubiquitination of substrate proteins [13]. The crystal structure of USP54 in complex with K63-linked diubiquitin reveals specialized binding interfaces that underlie this specificity, providing insights for engineering novel linkage-specific reagents.
Research Reagent Solutions for Ubiquitin Enrichment
The experimental toolkit for ubiquitin research has expanded significantly with the development of specialized reagents and methodologies. The following table summarizes key reagents for linkage-specific ubiquitin studies.
Table 2: Essential Research Reagents for Linkage-Specific Ubiquitin Studies
| Reagent Category | Specific Examples | Function & Application | Considerations |
|---|---|---|---|
| High-Affinity UBDs | OtUBD resin [17] [29] | Enrichment of mono- and polyubiquitinated proteins from lysates | More efficient for monoubiquitination than TUBEs; works with non-canonical sites |
| Deubiquitinase Inhibitors | N-ethylmaleimide (NEM), Chloroacetamide (CAA) [26] | Prevent ubiquitin chain disassembly during purification | NEM more potent but has more off-target effects; CAA more cysteine-specific |
| Linkage-Specific DUBs | OTUB1 (K48-specific), AMSH (K63-specific) [26] | Linkage verification (UbiCRest assay), chain editing | Used for validating chain linkage composition in samples |
| Specialized Enzymes | Ubc1 (K48-branching on K63 chains) [26] | Synthesis of defined branched ubiquitin chains | Enables production of complex ubiquitin architectures for binding studies |
| Chemical Tools | Propargylamide (PA)-based ubiquitin probes [13] | Activity profiling of DUBs, structural analysis | Forms vinyl thioether with catalytic cysteines of active DUBs |
| Chain Synthesis Systems | E2 enzyme combinations (CDC34-K48, Ubc13/Uev1a-K63) [26] | Production of defined linkage ubiquitin chains | Enzymatic synthesis preserves native isopeptide bonds |
The choice of deubiquitinase (DUB) inhibitors significantly impacts ubiquitin enrichment outcomes, as demonstrated in comparative studies of N-ethylmaleimide (NEM) and chloroacetamide (CAA) [26]. While NEM provides more complete inhibition of chain disassembly, it also exhibits greater off-target effects through alkylation of non-DUB cysteine residues and side reactions with N-termini and lysine side chains [26] [29]. CAA offers superior cysteine specificity but permits partial disassembly of Ub3 to Ub2 chains during pulldown experiments [26]. Researchers must therefore select inhibitors based on their specific experimental needs, balancing chain stability against potential perturbation of ubiquitin-binding surfaces.
Experimental Protocols for Linkage-Specific Ubiquitin Enrichment
OtUBD-Based Affinity Purification of Ubiquitinated Proteins
The OtUBD affinity resin provides a versatile platform for both native and denaturing purification of ubiquitinated proteins, accommodating diverse experimental requirements from interactome studies to direct ubiquitinome profiling [17] [29].
Reagents and Equipment:
- pRT498-OtUBD or pET21a-cys-His6-OtUBD plasmids (Addgene #190089, #190091)
- SulfoLink coupling resin
- Lysis buffers: Native (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP-40, protease inhibitors) or Denaturing (6 M guanidinium-HCl, 100 mM NaH₂PO₄, 10 mM Tris-HCl pH 8.0, 10 mM imidazole)
- Elution buffers: Competitor-based (10 mM reduced glutathione) or Denaturing (200 mM imidazole or 8 M urea)
- DUB inhibitors: N-ethylmaleimide (NEM) or chloroacetamide (CAA)
- Chromatography system or batch binding capability
Procedure:
- OtUBD Resin Preparation: Express and purify recombinant OtUBD protein from E. coli using standard protocols. Couple to SulfoLink resin via cysteine linkage according to manufacturer's instructions. Confirm coupling efficiency by Bradford assay or SDS-PAGE analysis [17].
Cell Lysis and Inhibitor Addition:
Affinity Purification:
- Incubate clarified lysate with OtUBD resin for 1-2 hours at 4°C with gentle rotation.
- Wash resin extensively with appropriate buffer (native or denaturing) containing reduced concentrations of DUB inhibitors.
- Elute bound proteins using either competitor-based elution (glutathione) or denaturing conditions (imidazole/urea) [17].
Downstream Analysis:
Critical Considerations: The choice between native and denaturing conditions determines which populations are isolated. Native lysis preserves non-covalent interactions between ubiquitinated proteins and their binding partners, enabling interactome studies. Denaturing conditions ensure isolation of only covalently ubiquitinated proteins for definitive ubiquitinome mapping [17] [29].
Linkage-Specific Ubiquitin Interactor Pulldown
This protocol enables identification of proteins that specifically recognize distinct ubiquitin chain architectures, using defined ubiquitin chains as bait [26].
Reagents and Equipment:
- Defined ubiquitin chains (mono-Ub, K48-Ub2, K48-Ub3, K63-Ub2, K63-Ub3, Br-Ub3)
- Streptavidin resin and biotinylation reagents
- Cell lysate (HeLa, yeast, or other relevant system)
- DUB inhibitors: NEM (5 mM) or CAA (20 mM)
- Crosslinker (DMP or DSS) optional
- Liquid chromatography-mass spectrometry (LC-MS/MS) system
Procedure:
- Ubiquitin Chain Preparation and Immobilization:
- Synthesize defined ubiquitin chains enzymatically using linkage-specific E2 enzymes (CDC34 for K48, Ubc13/Uev1a for K63, Ubc1 for branched chains) [26].
- Add serine/glycine linker with single cysteine residue to proximal Ub C-terminus.
- Conjugate biotin molecule using cysteine-maleimide chemistry. Verify complete biotinylation by intact MS [26].
- Immobilize biotinylated ubiquitin chains on streptavidin resin.
Pulldown Experiment:
- Prepare cell lysate in appropriate buffer supplemented with selected DUB inhibitor (NEM or CAA).
- Incubate lysate with ubiquitin-chain-conjugated resin for 1-2 hours at 4°C.
- Wash resin extensively with lysis buffer containing DUB inhibitors.
- Elute bound proteins with SDS-PAGE sample buffer or mild acid/competitor elution.
Analysis:
- Identify eluted proteins by LC-MS/MS.
- Use statistical comparison to identify proteins specifically enriched on particular chain types.
- Validate specific interactors by Western blotting or surface plasmon resonance (SPR) [26].
Methodological Considerations: The choice between NEM and CAA as DUB inhibitors will affect results. NEM provides nearly complete chain stabilization but may cause more off-target effects, while CAA is more specific but permits partial chain disassembly [26]. Including chains of different lengths (Ub2 vs Ub3) enables identification of length-dependent interactors, while branched chains reveal branch-specific binders.
The experimental workflow for linkage-specific ubiquitin interactor analysis is systematically outlined below, illustrating key stages from reagent preparation to data validation:
Figure 2: Linkage-Specific Ubiquitin Interactor Pulldown Workflow. The process encompasses defined ubiquitin chain preparation, immobilization, affinity pulldown with DUB inhibitors, and interactor identification with validation.
UbiCRest Linkage Verification Assay
The UbiCRest assay provides methodological validation of ubiquitin chain linkage composition using linkage-specific deubiquitinases, serving as an essential control for linkage-specific studies [26].
Reagents:
- OTUB1 (K48-specific DUB)
- AMSH (K63-specific DUB)
- Other linkage-specific DUBs as needed
- Ubiquitin chains or ubiquitinated samples
- Reaction buffer (50 mM Tris-HCl pH 7.5, 50 mM NaCl, 1 mM DTT, 5 mM MgCl₂)
Procedure:
- Divide ubiquitin chain sample into aliquots for each DUB treatment and no-enzyme control.
- Incubate with respective linkage-specific DUBs (OTUB1 for K48, AMSH for K63) for 1-2 hours at 37°C.
- Analyze digestion products by SDS-PAGE and Western blotting with anti-ubiquitin antibody.
- Interpret linkage composition based on digestion pattern: Complete digestion indicates homotypic chains of that linkage, partial digestion suggests mixed or branched chains [26].
Applications and Emerging Research Directions
Linkage-specific UBDs have enabled significant advances in understanding ubiquitin-dependent processes, particularly in characterizing the functions of atypical ubiquitin chains and branched ubiquitin architectures. Recent research has revealed that K6-linked chains play crucial roles in mitophagy and DNA damage response, with E3 ligases like Parkin and HUWE1 assembling K6-linked chains during mitochondrial quality control and genomic stress responses [28]. K11-linked chains have emerged as important regulators of cell cycle progression, particularly through the anaphase-promoting complex/cyclosome (APC/C) that constructs K11/K48-branched chains to enhance substrate degradation during mitosis [28]. K29- and K33-linked chains participate in non-degradative functions including kinase regulation and trafficking, while K27-linked chains contribute to immune signaling pathways [28].
Branched ubiquitin chains represent a frontier in ubiquitin research, with K48/K63-branched chains constituting approximately 20% of cellular K63 linkages and serving dual functions in enhancing proteasomal degradation and modulating NF-κB signaling [26] [3]. The development of enzymatic methods for synthesizing defined branched ubiquitin chains using engineered E2 enzymes like Ubc1 has enabled systematic investigation of these complex architectures [26] [3]. These tools have revealed that proteins including PARP10, UBR4, and huntingtin-interacting protein HIP1 exhibit specific binding to K48/K63-branched ubiquitin chains, suggesting specialized recognition mechanisms for these hybrid architectures [26].
Emerging technologies continue to expand the toolkit for ubiquitin research. Genetic code expansion approaches enable site-specific incorporation of non-canonical amino acids for precise ubiquitin chain assembly, while chemical synthesis methods permit construction of ubiquitin chains with defined linkages and strategically placed modifications [3]. Photo-controlled enzymatic assembly using photolabile NVOC-protected lysine residues enables sequential construction of complex ubiquitin architectures [3]. These advanced methodologies, combined with linkage-specific UBDs, promise to accelerate our decoding of the ubiquitin code and its multifaceted roles in cellular regulation and disease pathogenesis.
Linkage-specific UBDs represent indispensable tools for deciphering the complex language of ubiquitin signaling, enabling researchers to isolate and characterize specific ubiquitin chain architectures from complex biological mixtures. The continuing development of novel affinity reagents like OtUBD, alongside the refinement of traditional methodologies, has significantly enhanced our capacity to map ubiquitin-modified proteins and their interaction networks. As research extends beyond the canonical K48 and K63 linkages to encompass the full spectrum of atypical ubiquitin chains and branched architectures, these tools will play increasingly critical roles in elucidating the physiological functions of diverse ubiquitin signals. The integration of linkage-specific UBDs with advanced proteomic, structural, and chemical biology approaches promises to unlock new dimensions of the ubiquitin code, with profound implications for understanding cellular regulation and developing targeted therapeutic interventions for ubiquitin-related diseases.
Protein ubiquitylation is a crucial post-translational modification regulating diverse cellular processes, including protein degradation, DNA repair, and immune signaling [29] [30]. The ubiquitin-proteasome system (UPS) serves as the core machinery for targeted protein degradation and quality control in eukaryotes, playing a pivotal role in maintaining proteostasis and cellular homeostasis [1]. Defects in ubiquitylation are connected to many human disorders, such as cancers, viral infections, and neurodegenerative diseases, making the development of sensitive methods to study the ubiquitylated proteome critically important [29] [30].
Ubiquitin-binding domains (UBDs) have emerged as powerful tools for detecting and purifying ubiquitylated proteins. This application note details two specialized strategies: the use of the ZnF_UBP domain from USP5 for purifying endogenous unanchored polyubiquitin chains (FUBEs), and the high-affinity OtUBD from Orientia tsutsugamushi for versatile enrichment of diverse ubiquitylated substrates.
OtUBD: A Versatile Tool for Ubiquitinome Profiling
The OtUBD is a high-affinity ubiquitin-binding domain derived from a deubiquitylase (DUB) effector protein produced by the intracellular bacterium Orientia tsutsugamushi [29] [25]. This UBD binds monomeric ubiquitin at the isoleucine-44 hydrophobic patch with an unprecedented dissociation constant (Kd) of approximately 5 nM, which is more than 500-fold tighter than any other natural UBD described to date [25]. The co-crystal structure of OtDUB with ubiquitin revealed three bound ubiquitins: one engages the S1 site, the second binds an S2 site contributing to chain specificity, and the third binds a unique UBD [25].
Table 1: Key Characteristics of OtUBD
| Parameter | Specification |
|---|---|
| Source | Orientia tsutsugamushi OtDUB (residues 170-264) |
| Affinity for Monoubiquitin | ~5 nM Kd [25] |
| Binding Site | Isoleucine-44 hydrophobic patch of ubiquitin [25] |
| Recognized Ubiquitin Modifications | MonoUb, PolyUb (all linkages), non-canonical ubiquitylation [4] [29] |
| Compatible Samples | Baker's yeast, mammalian cell lysates, other biological samples [4] |
| Downstream Applications | Immunoblotting, LC-MS/MS proteomics, UbiCREST [4] |
Experimental Protocol for OtUBD-Based Enrichment
Materials and Reagents
- Recombinant OtUBD (His6-tagged or MBP-fused)
- Affinity resin: Amylose resin for MBP-tagged fusions, Ni-NTA for His6-tagged fusions
- Lysis buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, protease inhibitors (e.g., PMSF), DUB inhibitors (e.g., NEM)
- Wash buffers: Native (for interactome) and Denaturing (for ubiquitinome)
- Elution buffer: SDS-PAGE sample buffer or competitive elution with free ubiquitin
Procedure
Cell Lysis and Preparation: Lyse cells in appropriate buffer supplemented with DUB inhibitors (NEM) to prevent deubiquitylation. For yeast cells, mechanical disruption is recommended; for mammalian cells, detergent-based lysis is sufficient [4] [30].
Resin Preparation: Immobilize recombinant MBP-OtUBD or MBP-3xOtUBD on amylose resin. For His6-tagged OtUBD, use Ni-NTA resin. Wash resin extensively with lysis buffer before use [30].
Pulldown Incubation: Incubate clarified cell lysate with OtUBD-bound resin for 1-2 hours at 4°C with gentle rotation. The recommended amount is 3 μM OtUBD for protection assays [30].
Washing: Based on experimental goals:
- Native Conditions: Use mild wash buffer (e.g., 50 mM Tris-HCl pH 7.5, 150 mM NaCl) to co-purify both covalently ubiquitylated proteins and their interacting partners [4].
- Denaturing Conditions: Use high-stringency buffer (e.g., with 1% SDS or 2 M urea) to selectively enrich covalently ubiquitylated proteins only [4].
Elution: Elute bound proteins with SDS-PAGE sample buffer by heating at 95°C for 5-10 minutes, or competitively elute with free ubiquitin (1-2 mg/mL) [4] [30].
Downstream Analysis: Process eluates for immunoblotting with anti-ubiquitin antibodies or for LC-MS/MS proteomic analysis to profile the ubiquitylome [4].
Figure 1: OtUBD Experimental Workflow. The protocol offers both native and denaturing pathways to selectively study the ubiquitin interactome or covalently ubiquitinated proteins, respectively.
Key Advantages and Applications
- Broad Specificity: Efficiently enriches both monoubiquitylated and polyubiquitylated proteins, unlike TUBEs which have low affinity for monoubiquitylation [29] [30].
- Linkage-Unbiased Recognition: Capable of detecting all ubiquitin chain linkage types without significant bias [29].
- Non-Canonical Ubiquitylation Detection: Can identify ubiquitin linkages to non-lysine residues (serine, threonine, cysteine) and N-terminal ubiquitylation [29].
- Protection Function: When added to cell lysates, OtUBD protects ubiquitylated proteins from degradation by endogenous DUBs, similar to NEM treatment [30].
- Proteomic Discovery: Combined with quantitative proteomics, OtUBD has identified potential substrates for E3 ligases Bre1 and Pib1 in budding yeast [29] [30].
FUBEs: Specific Purification of Unanchored Polyubiquitin Chains
Unanchored polyubiquitin chains are endogenous non-substrate linked ubiquitin polymers with emerging roles in cellular physiology, particularly in signaling pathways [31]. Recent research has highlighted the importance of unconventional K29-linked unanchored polyubiquitin chains in affecting ribosome biogenesis and directing ribosomal proteins to the intranuclear quality control compartment [32]. The method for purifying these endogenous unanchored polyubiquitin chains utilizes the ZnF_UBP domain of the deubiquitinating enzyme USP5, which selectively binds unanchored polyubiquitin chains with high specificity [31].
Table 2: Key Characteristics of FUBE (USP5 ZnF_UBP) Method
| Parameter | Specification |
|---|---|
| Source | Human USP5 deubiquitinating enzyme |
| Target | Endogenous unanchored polyubiquitin chains |
| Specificity | Selective for free polyubiquitin chains over substrate-linked chains [31] |
| Chain Type Recognition | Multiple linkage types, including K29-linked [32] |
| Key Applications | Study of unanchored chain biology, signaling pathways |
Experimental Protocol for FUBE-Based Purification
Materials and Reagents
- Recombinant ZnF_UBP domain of USP5
- Affinity resin appropriate for tag (GST, His, etc.)
- Lysis buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5% NP-40, protease inhibitors, DUB inhibitors (NEM)
- Wash buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% NP-40
- Elution buffer: Competitive elution with free ubiquitin or SDS-PAGE buffer
Procedure
Cell Lysis: Lyse cells in appropriate buffer containing DUB inhibitors (NEM) to preserve unanchored chains. Use gentle detergent conditions to maintain protein complexes if needed [31].
Affinity Resin Preparation: Immobilize recombinant ZnF_UBP domain on appropriate affinity resin based on tag. Wash thoroughly with lysis buffer before use.
Binding Incubation: Incubate clarified cell lysate with ZnF_UBP-bound resin for 1-2 hours at 4°C with gentle rotation [31].
Washing: Wash resin extensively with wash buffer to remove non-specifically bound proteins. The mild detergent concentration helps maintain specificity while removing contaminants.
Elution: Elute bound unanchored polyubiquitin chains competitively using free ubiquitin (0.5-1 mg/mL) or with SDS-PAGE sample buffer for direct analysis [31].
Detection and Analysis: Analyze eluates by immunoblotting with anti-ubiquitin antibodies or subject to molecular analysis for chain length and linkage type determination [31].
Figure 2: FUBE Purification Workflow. The ZnF_UBP domain of USP5 selectively purifies endogenous unanchored polyubiquitin chains through affinity binding and competitive elution.
Key Advantages and Applications
- Specificity for Unanchored Chains: Selective purification of free polyubiquitin chains without contamination from substrate-linked ubiquitin [31].
- Endogenous Material: Works with endogenous ubiquitin levels, avoiding potential artifacts from tagged ubiquitin overexpression [31].
- Functional Studies: Enables investigation of unanchored chain functions in signaling pathways and cellular physiology [31] [32].
- Linkage Diversity: Can purify various linkage types of unanchored chains, including the less common K29-linked chains that function in ribosome biogenesis and intranuclear quality control [32].
Comparative Analysis and Implementation Strategy
Side-by-Side Comparison
Table 3: Strategic Selection Guide: OtUBD vs. FUBE Applications
| Parameter | OtUBD | FUBE (ZnF_UBP) |
|---|---|---|
| Primary Target | Covalently ubiquitylated substrates (mono & poly) | Unanchored/free polyubiquitin chains |
| Specificity | Broad (entire ubiquitinome) | Narrow (unanchored chains only) |
| Affinity | ~5 nM Kd for monoubiquitin [25] | Not specified in sources |
| Monoubiquitin Detection | Excellent [30] | Not applicable |
| Polyubiquitin Chain Detection | All linkage types, unbiased [29] | Multiple linkages including K29 [32] |
| Non-Canonical Sites | Detects serine, threonine, cysteine ubiquitylation [29] | Not specified |
| Best Applications | Ubiquitylome profiling, substrate identification, interaction studies | Unanchored chain biology, specialized signaling pathways |
Research Reagent Solutions
Table 4: Essential Research Reagents for UBD-Based Affinity Enrichment
| Reagent | Function | Examples/Specifications |
|---|---|---|
| Recombinant OtUBD | High-affinity ubiquitin binding | His6-tagged or MBP-fused; 3xOtUBD for increased capacity [30] |
| ZnF_UBP Domain (USP5) | Unanchored chain purification | GST or His-tagged recombinant protein [31] |
| DUB Inhibitors | Preserve ubiquitylated species | N-ethylmaleimide (NEM), PR-619 [4] [30] |
| Protease Inhibitors | Prevent protein degradation | PMSF, complete protease inhibitor cocktails [4] |
| Affinity Resins | Immobilization of UBDs | Amylose resin (MBP-tag), Ni-NTA (His-tag), Glutathione resin (GST-tag) [4] [30] |
| Anti-Ubiquitin Antibodies | Detection of enriched ubiquitin | Linkage-specific (e.g., anti-K48, anti-K63) or pan-ubiquitin (e.g., FK1, FK2) [29] |
Technical Considerations and Troubleshooting
Optimization Guidelines
- Inhibition Efficiency: Always include effective DUB inhibitors (NEM at 5-10 mM) during cell lysis to prevent loss of ubiquitylated species through deubiquitylation [4] [30].
- Buffer Compatibility: For OtUBD interactome studies, use native wash conditions; for specific ubiquitinome analysis, implement denaturing washes to remove non-covalently bound interactors [4].
- Capacity Considerations: For samples rich in ubiquitylated proteins, use the MBP-3xOtUBD construct instead of single OtUBD for increased binding capacity [30].
- Control Experiments: Include appropriate controls such as resin-only samples and competition with free ubiquitin to confirm specificity of enrichment.
Advanced Applications
- Quantitative Proteomics: Combine OtUBD enrichment with SILAC, TMT, or label-free quantitation for comprehensive ubiquitylome profiling [4] [29].
- UbiCREST Analysis: Use OtUBD-enriched material with linkage-specific DUBs to determine ubiquitin chain topology [4].
- Interaction Studies: Employ crosslinking after OtUBD enrichment under native conditions to stabilize transient ubiquitin-dependent interactions.
- High-Throughput Screening: Recent advancements have led to development of ThUBD-coated 96-well plates for high-throughput detection of ubiquitination signals, demonstrating the adaptability of UBD-based methods to screening platforms [1].
The specialized strategies presented here—FUBEs for unanchored polyubiquitin chains and OtUBD for versatile ubiquitinome purification—provide researchers with powerful tools for dissecting the complexity of ubiquitin signaling. The ZnF_UBP-based method offers specificity for studying the emerging roles of unanchored chains in cellular physiology, while OtUBD delivers unprecedented versatility in capturing diverse ubiquitylation events, including monoubiquitylation, polyubiquitylation of all linkages, and non-canonical ubiquitylation. Implementation of these methods enables comprehensive analysis of ubiquitin-dependent processes in health and disease, supporting drug discovery efforts targeting the ubiquitin-proteasome system.
Protein ubiquitination is a crucial post-translational modification that regulates diverse cellular processes, including proteasomal degradation, DNA repair, and signal transduction [19] [27]. The study of ubiquitinated proteins is challenging due to their low abundance and the complexity of ubiquitin chain architectures. Affinity enrichment methods are essential for isolating these modifications for downstream analysis. This protocol details the use of the OtUBD affinity resin, a high-affinity ubiquitin-binding domain derived from Orientia tsutsugamushi, for the enrichment of ubiquitinated proteins from both baker's yeast and mammalian cell lysates [17] [4]. The following sections provide a comprehensive guide for researchers to perform these enrichments under either denaturing conditions, which specifically isolate covalently ubiquitinated proteins (the ubiquitinome), or native conditions, which also co-purify ubiquitin- or ubiquitinated protein-interacting proteins (the ubiquitin interactome) [17] [29].
Key Features of the OtUBD Method
The OtUBD-based affinity enrichment offers several distinct advantages over other methods like tandem ubiquitin-binding entities (TUBEs) or antibody-based immunoprecipitation [29] [19].
Table 1: Comparison of Ubiquitinated Protein Enrichment Methods
| Method | Key Features | Advantages | Limitations |
|---|---|---|---|
| OtUBD Affinity Resin | High-affinity UBD; Enriches mono- and polyubiquitinated proteins [17] | Versatile (works in denaturing/native conditions); Cost-effective; Detects non-lysine ubiquitination [29] | Requires preparation of recombinant OtUBD and resin |
| TUBEs | Multiple low-affinity UBDs in tandem [19] | Protects polyubiquitin chains from deubiquitinases (DUBs) [19] | Poor efficiency for monoubiquitinated proteins [29] [19] |
| Antibody-based (e.g., P4D1, FK2) | Antibodies recognizing ubiquitin epitopes [19] | Does not require genetic manipulation (works at endogenous levels) [19] | High cost; Potential for non-specific binding; May lack sensitivity [17] [19] |
| DiGly Antibody (K-ε-GG) | Antibodies recognizing diglycine remnant on lysine after trypsin digestion [29] [19] | Excellent for proteomic identification of ubiquitination sites on lysine [19] | Cannot identify non-lysine ubiquitination (e.g., serine, threonine, cysteine) [29] |
The Scientist's Toolkit: Essential Reagents and Materials
The following table lists the key reagents and materials required to successfully perform the OtUBD enrichment protocol.
Table 2: Essential Research Reagent Solutions
| Item | Function / Application | Examples / Specifications |
|---|---|---|
| OtUBD Plasmids | Source for recombinant OtUBD protein production [17] | pRT498-OtUBD (Addgene #190089); pET21a-cys-His6-OtUBD (Addgene #190091) [17] |
| Affinity Resin | Solid support for coupling OtUBD and affinity purification | SulfoLink Coupling Resin (for covalent coupling) [17] |
| Cell Lysis Buffers | Solubilizing proteins from cells while preserving ubiquitin modifications | Denaturing: SDS-containing buffer [17]. Native: Triton-X-100 or NP-40 based buffer [17] |
| Protease Inhibitors | Prevent protein degradation during lysis and purification | cOmplete EDTA-free protease inhibitor cocktail [17] |
| Deubiquitinase (DUB) Inhibitors | Preserve ubiquitin conjugates by inhibiting DUB activity | N-ethylmaleimide (NEM) [17] |
| Reducing Agents | Maintain reduced cysteine residues and protein stability | Dithiothreitol (DTT) or Tris(2-carboxyethyl)phosphine (TCEP) [17] |
| Elution Buffers | Release bound ubiquitinated proteins from the OtUBD resin | SDS-PAGE sample buffer (denaturing) or a buffer with high concentration of free ubiquitin (native) [17] |
| Primary Antibodies | Detect ubiquitinated proteins via immunoblotting | Anti-ubiquitin mouse mAb P4D1 (1:1,000-1:4,000); Anti-ubiquitin rabbit Ab (E412J) (1:4,000) [17] |
Detailed Step-by-Step Protocol
A. Preparation of OtUBD Affinity Resin
This section outlines the production of the key reagent, the OtUBD affinity resin.
- Protein Expression: Transform the pET21a-cys-His6-OtUBD plasmid into an appropriate E. coli expression strain. Induce protein expression with Isopropyl β-D-1-thiogalactopyranoside (IPTG) [17].
- Protein Purification: Lyse the bacterial cells and purify the recombinant Cys-His6-OtUBD protein using Immobilized Metal Affinity Chromatography (IMAC) with Ni-NTA agarose, taking advantage of the hexahistidine tag [17].
- Resin Coupling: Conjugate the purified OtUBD protein to the SulfoLink coupling resin via its cysteine residue, following the manufacturer's instructions. The result is a stable, high-density OtUBD affinity resin [17].
- Storage: Resuspend the prepared resin in a storage buffer (e.g., PBS with 0.02% sodium azide) and store at 4°C.
B. Cell Lysis and Lysate Preparation
The lysis conditions are critical and must be chosen based on the experimental goal.
- Cell Harvesting: Collect yeast or mammalian cells by centrifugation. Wash cells with cold PBS [17].
- Lysis Buffer Formulation:
- For Denaturing Conditions (RIPA-SDS-DTT Buffer): Use a lysis buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 1% SDS, 1 mM EDTA, and 1 mM DTT. The strong denaturant SDS and high concentration of DTT disrupt non-covalent protein-protein interactions, ensuring that only covalently ubiquitinated proteins are isolated [17].
- For Native Conditions (RIPA Buffer): Use a lysis buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA, and 1 mM DTT. The milder detergents preserve native interactions, allowing for the co-purification of ubiquitinated proteins and their interacting partners [17].
- Inhibitors: Supplement both lysis buffers with fresh protease inhibitors (e.g., PMSF or commercial cocktails) and 10 mM N-ethylmaleimide (NEM) to inhibit deubiquitinases [17].
- Lysis: Lyse the cells by vortexing with glass beads (for yeast) or by sonication/passage through a needle (for mammalian cells). Clarify the lysate by centrifugation at high speed (e.g., 16,000 × g for 15 min) [17].
- Protein Quantification: Determine the protein concentration of the supernatant using a compatible assay (e.g., Pierce BCA or Bradford assay) [17].
C. OtUBD Affinity Pulldown
- Equilibration: Equilibrate the OtUBD affinity resin in the corresponding lysis buffer (without SDS for native conditions).
- Incubation: Incub the clarified cell lysate with the equilibrated OtUBD resin for 2-4 hours at 4°C with gentle rotation.
- Washing:
- For Denaturing Conditions: Wash the resin 3-4 times with a wash buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 1% SDS, 1 mM EDTA, and 1 mM DTT [17].
- For Native Conditions: Wash the resin 3-4 times with a wash buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA, and 1 mM DTT [17].
D. Elution of Bound Proteins
Elution methods can be selected based on the desired downstream application.
- Elution for Immunoblotting: For direct detection by SDS-PAGE and western blotting, elute the bound proteins by boiling the resin in 1X SDS-PAGE sample buffer for 5-10 minutes [17].
- Competitive Elution for Protein Recovery: For applications requiring functional proteins (e.g., proteomics), elute by incubating the resin with a buffer containing 1-2 M urea or a high concentration (e.g., 1 mg/mL) of free ubiquitin, which competes with the ubiquitinated proteins for binding to OtUBD [17].
The experimental workflow for both denaturing and native conditions is summarized in the diagram below.
Expected Results and Downstream Applications
Successful execution of this protocol will yield enriched ubiquitinated proteins suitable for various analyses. When analyzed by immunoblotting with anti-ubiquitin antibodies, a characteristic high-molecular-weight smear is typically observed, representing the diverse population of mono- and polyubiquitinated proteins [17] [29]. The denaturing protocol will yield a cleaner smear of covalently modified proteins, while the native protocol may show additional specific bands corresponding to interactors.
For proteomic applications, the eluted proteins can be digested with trypsin and analyzed by Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS). Combining data from denaturing and native pulldowns helps distinguish direct ubiquitination targets from mere interacting proteins [17] [4]. This approach has been successfully used to identify potential substrates for specific E3 ligases, such as Bre1 and Pib1 in budding yeast [29].
Troubleshooting Guide
Table 3: Common Issues and Potential Solutions
| Problem | Potential Cause | Suggested Solution |
|---|---|---|
| High background / non-specific binding | Incomplete washing or non-optimal resin | Increase number of washes; include 500 mM NaCl in wash buffers to reduce non-specific ionic interactions; ensure proper preparation and blocking of the OtUBD resin. |
| Low yield of ubiquitinated proteins | Inefficient lysis, DUB activity, or weak binding | Ensure complete cell disruption; always include DUB inhibitors (NEM) in lysis and wash buffers; verify the activity and coupling efficiency of the OtUBD resin. |
| No signal in western blot | Ubiquitinated proteins are too dilute | Concentrate the eluted sample (e.g., by TCA precipitation) before loading the gel; increase the amount of starting lysate. |
| Distinct bands instead of a smear | Potential degradation or highly specific enrichment | Ensure adequate levels of protease inhibitors; this can also be an expected result if studying a specific, highly enriched ubiquitinated protein. |
The OtUBD affinity enrichment protocol provides a versatile, powerful, and economical method for probing the ubiquitinome and ubiquitin interactome. Its ability to function robustly under both denaturing and native conditions, coupled with its high affinity for both mono- and polyubiquitin conjugates, offers distinct advantages over other methodologies [17] [29]. By following this detailed step-by-step guide, researchers can reliably isolate ubiquitinated proteins to advance our understanding of the complex ubiquitin code in health and disease.
Ubiquitin Binding Domain (UBD)-based affinity enrichment has emerged as a powerful and versatile strategy for investigating the ubiquitin code. Unlike methods that rely on tagged ubiquitin overexpression, UBD-based approaches allow for the study of endogenous ubiquitination under physiological conditions, providing a more accurate representation of cellular signaling [19]. The OtUBD (Orientia tsutsugamushi Ubiquitin-Binding Domain), in particular, is a high-affinity UBD with a dissociation constant in the low nanomolar range, making it highly effective for enriching both mono- and poly-ubiquitinated proteins from complex biological samples [17] [4]. This application note details standardized protocols for coupling OtUBD-based enrichment with three critical downstream applications: immunoblotting, proteomics, and UbiCREST. These integrated workflows enable researchers to detect ubiquitinated proteins, identify specific ubiquitination sites and interacting proteins on a systems-wide scale, and decipher ubiquitin chain linkage types, respectively.
Key Applications and Workflows
The table below summarizes the primary applications, key characteristics, and recommended workflows for UBD-based enrichment.
Table 1: Overview of Downstream Applications for UBD-Based Enrichment
| Application | Key Objective | Recommended UBD Workflow | Key Outcome Measures |
|---|---|---|---|
| Immunoblotting | Detect and confirm protein ubiquitination | Denaturing or Native | Visualization of ubiquitin smears/specific bands via anti-ubiquitin antibodies. |
| Proteomics (LC-MS/MS) | Identify ubiquitination sites & interacting proteins | Denaturing (Ubiquitinome) or Native (Interactome) | Large-scale identification and quantification of ubiquitinated peptides/proteins. |
| UbiCREST | Determine ubiquitin chain linkage topology | Native (followed by deubiquitinase assay) | Linkage-specific cleavage patterns revealed by immunoblotting. |
Detailed Experimental Protocols
UBD Enrichment Coupled with Immunoblotting
Immunoblotting is a fundamental method for validating protein ubiquitination. The OtUBD enrichment enhances the signal-to-noise ratio by concentrating ubiquitinated species prior to analysis.
Protocol:
- Cell Lysis (Denaturing): For direct ubiquitinated protein analysis, lyse cells in a denaturing buffer (e.g., 1% SDS, 50 mM Tris-HCl pH 7.5, 5 mM N-ethylmaleimide (NEM), 1 mM phenylmethylsulfonyl fluoride (PMSF), and cOmplete EDTA-free protease inhibitor cocktail). Immediately boil lysates for 10 minutes to inactivate deubiquitinases (DUBs) [17].
- Lysate Preparation: Dilute the denatured lysate 10-fold with a non-denaturing buffer (e.g., 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100) to reduce SDS concentration for compatibility with the OtUBD resin.
- UBD Pulldown: Incubate the clarified lysate with OtUBD affinity resin for 2 hours at 4°C with gentle agitation.
- Washing: Pellet the resin and wash extensively with wash buffer (e.g., 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1% Triton X-100) to remove non-specifically bound proteins.
- Elution: Elute bound ubiquitinated proteins by boiling the resin in 1X Laemmli SDS-PAGE sample buffer containing 50 mM DTT for 5-10 minutes.
- Immunoblotting: Resolve eluates by SDS-PAGE and transfer to a PVDF membrane. Probe with anti-ubiquitin antibodies (e.g., P4D1 at 1:1,000 or E412J at 1:4,000) and corresponding HRP-conjugated secondary antibodies for detection [17].
UBD Enrichment Coupled with Proteomics
Coupling OtUBD enrichment with liquid chromatography-tandem mass spectrometry (LC-MS/MS) enables systems-level analysis of the ubiquitinome and ubiquitin interactome.
Protocol:
- Experimental Design:
- For Ubiquitinome Analysis: Use the denaturing workflow (Section 3.1, steps 1-5) to exclusively isolate covalently ubiquitinated proteins.
- For Interactome Analysis: Use a native lysis buffer (e.g., 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, supplemented with NEM and protease inhibitors) to preserve non-covalent interactions between ubiquitinated proteins and their binding partners [17] [4].
- Trypsin Digestion: After UBD enrichment and elution, reduce and alkylate proteins. Digest the protein mixture with sequencing-grade trypsin.
- Peptide Cleanup: Desalt the resulting peptides using C18 solid-phase extraction tips or columns.
- LC-MS/MS Analysis:
- Data-Dependent Acquisition (DDA): Suitable for building spectral libraries. Fractionating peptides prior to MS analysis can significantly deepen coverage [33].
- Data-Independent Acquisition (DIA): Recommended for superior quantitative accuracy and data completeness. A tailored DIA method with 46 precursor isolation windows and a fragment scan resolution of 30,000 has been optimized for diGly-peptide analysis [33].
- Data Analysis: Search MS/MS spectra against a relevant protein database. Identify ubiquitination sites by searching for the diGly remnant (K-ε-GG), a tryptic signature of ubiquitination, with a mass shift of +114.04 Da on modified lysines [33] [19].
UBD Enrichment in the UbiCREST Assay
The UbiCREST (Ubiquitin Chain Restriction) assay leverages linkage-specific deubiquitinases (DUBs) to decipher the topology of ubiquitin chains on enriched substrates.
Protocol:
- UBD Enrichment under Native Conditions: Enrich ubiquitinated proteins from native cell lysates using the OtUBD resin, as described in the interactome protocol (Section 3.2).
- Equilibration: Wash the resin with a reaction buffer compatible with DUB activity (e.g., 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM DTT).
- DUB Treatment: Split the resin into several aliquots. Incubate each aliquot with a recombinant linkage-specific DUB (e.g., OTUD1 for K48-linked chains, OTUB1 for K48/K11-linked chains, AMSH for K63-linked chains, etc.) or a control buffer for 1-2 hours at 37°C [17].
- Termination and Analysis: Stop the reaction by adding SDS-PAGE sample buffer and boiling. Analyze the cleavage products by immunoblotting with anti-ubiquitin antibodies or antibodies against the protein substrate of interest. The specific cleavage pattern reveals the chain linkages present.
The Scientist's Toolkit: Key Research Reagent Solutions
The table below catalogues essential reagents and their functions for implementing the described UBD-based workflows.
Table 2: Essential Research Reagents for UBD-Based Applications
| Reagent / Tool | Function / Application | Examples / Specifications |
|---|---|---|
| OtUBD Affinity Resin | High-affinity capture of mono- and poly-ubiquitinated proteins from lysates. | Resin prepared using recombinant cys-His6-OtUBD (Addgene #190091) coupled to SulfoLink resin [17]. |
| Ubiquitin Antibodies | Detection of ubiquitinated proteins in immunoblotting. | P4D1 (mouse monoclonal, 1:1,000), E412J (rabbit monoclonal, 1:4,000) [17]. |
| Protease Inhibitors | Prevent proteolytic degradation of ubiquitin conjugates during isolation. | cOmplete EDTA-free protease inhibitor cocktail; 1 mM PMSF [17]. |
| Deubiquitinase (DUB) Inhibitors | Preserve ubiquitin signals by preventing chain disassembly by endogenous DUBs. | 5-20 mM N-Ethylmaleimide (NEM) or 1-5 mM Iodoacetamide included in lysis buffer [17] [33]. |
| Linkage-Specific DUBs | Digest specific ubiquitin linkages in the UbiCREST assay. | OTUD1 (K48-specific), AMSH (K63-specific) [17]. |
| diGly Remnant Antibodies | Immunoaffinity enrichment of ubiquitinated peptides for MS-based proteomics. | PTMScan Ubiquitin Remnant Motif (K-ε-GG) Kit; used after tryptic digest [33]. |
Optimizing UBD Enrichment: Strategies for Enhanced Specificity and Yield
Protein ubiquitination represents a crucial post-translational modification that regulates diverse cellular functions, including protein degradation, DNA repair, and signal transduction [19]. However, the low stoichiometry of this modification—where ubiquitinated proteins often represent only a minute fraction of their total cellular counterparts—presents a fundamental analytical challenge. This low abundance, combined with the transient nature of ubiquitination events and the structural complexity of ubiquitin chains, necessitates highly efficient enrichment strategies to enable accurate detection and characterization [19]. The emergence of Ubiquitin-Binding Domain (UBD) based affinity enrichment methods has revolutionized this field by providing tools capable of selectively isolating these rare ubiquitination events from complex biological samples.
The versatility of ubiquitination further complicates its analysis. Ubiquitin can modify substrate proteins as a single moiety (monoubiquitination), multiple single moieties (multiple monoubiquitination), or polymers (polyubiquitination) forming chains through different linkage types—K48, K63, M1, and others—each encoding distinct functional outcomes [19] [34]. UBD-based technologies have evolved to address this complexity, enabling researchers to capture the full spectrum of ubiquitination events, which is essential for understanding ubiquitin signaling in both physiological and pathological contexts, including cancer and neurodegenerative diseases [19].
UBD-Based Enrichment Technologies: A Comparative Analysis
Several UBD-based affinity tools have been developed to address the challenges of low-stoichiometry ubiquitination. The following table provides a systematic comparison of the primary technologies currently advancing the field.
Table 1: Comparison of UBD-Based Affinity Enrichment Technologies
| Technology | Key Features | Affinity & Specificity | Applications | Limitations |
|---|---|---|---|---|
| OtUBD [4] [24] | High-affinity domain from Orientia tsutsugamushi; native and denaturing workflows | Low nanomolar range; enriches both mono- and polyubiquitinated proteins | Immunoblotting, proteomics, UbiCREST; yeast and mammalian systems | Requires recombinant protein purification |
| TUBEs (Tandem Ubiquitin Binding Entities) [35] [19] | Four tandem UBA domains from ubiquilin-1; semi-denaturing conditions | Broad specificity for polyubiquitin chains; low nanomolar affinity | Monitoring small molecule-induced ubiquitination changes; target validation | Lower affinity for monoubiquitination; potential linkage bias in some formulations |
| ThUBD (Tandem Hybrid Ubiquitin-Binding Domains) [1] | Engineered tandem hybrid domains; 96-well plate format | Unbiased recognition of all ubiquitin chain types; high affinity | High-throughput screening; PROTAC development; drug discovery | Specialized format less suitable for small-scale experiments |
Each technology offers distinct advantages depending on the research objectives. OtUBD provides exceptional versatility for both discovery and validation workflows, while TUBEs excel in preserving labile ubiquitination signals during extraction. ThUBD represents a significant advancement for high-throughput applications, particularly in drug discovery contexts where screening efficiency is paramount [1].
Table 2: Quantitative Performance Characteristics of UBD Technologies
| Technology | Enrichment Capacity | Detection Sensitivity | Compatible Input Amounts | Chain Type Coverage |
|---|---|---|---|---|
| OtUBD [4] | High for both mono- and polyUb | Sub-microgram scale | 1-10 mg lysate | All types |
| TUBEs [35] | Very high for polyUb | Nanogram scale for immunoblotting | 2-5 mg lysate | All major linkages (K48, K63 >90%) |
| ThUBD-coated plates [1] | ~5 pmol polyubiquitin chains | High-throughput (96-well format) | Variable sample volumes | Unbiased recognition |
Detailed Experimental Protocols
OtUBD-Based Enrichment for Low-Abundance Ubiquitinated Proteins
The OtUBD system represents one of the most robust methods for enriching low-stoichiometry ubiquitination events due to its exceptional affinity for ubiquitin [4] [24].
Materials and Reagents:
- pRT498-OtUBD or pET21a-cys-His6-OtUBD plasmid (Addgene #190089, #190091)
- SulfoLink coupling resin (Thermo Scientific, #20402)
- L-cysteine (Sigma-Aldrich, #C7352) for resin preparation
- Lysis Buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 1 mM EDTA, 10% glycerol
- Protease Inhibitor Cocktail (Roche, #11873580001)
- 20 mM N-ethylmaleimide (NEM) (Sigma-Aldrich, #E3876) to inhibit deubiquitinases
- DNase I (Roche, #10104159001)
- Elution Buffer: 50 mM Tris-HCl (pH 7.5), 2% SDS, 150 mM NaCl
Step-by-Step Protocol:
Recombinant OtUBD Purification:
- Transform pRT498-OtUBD into BL21(DE3) E. coli and culture in LB medium with appropriate antibiotics.
- Induce expression with 0.5 mM IPTG at 18°C for 16 hours.
- Pellet cells and resuspend in Lysis Buffer.
- Lyse cells by sonication and clarify lysate by centrifugation.
- Purify OtUBD using Ni-NTA agarose according to standard protocols.
OtUBD Affinity Resin Preparation:
- Reduce SulfoLink resin according to manufacturer's instructions.
- Couple purified OtUBD to resin via cysteine residues.
- Block remaining reactive groups with L-cysteine.
- Wash resin extensively and store in PBS with 0.02% sodium azide at 4°C.
Sample Preparation and Enrichment:
- Harvest cells and lyse in Lysis Buffer supplemented with protease inhibitors and 20 mM NEM.
- Clarify lysate by centrifugation at 20,000 × g for 15 minutes.
- Incubate 1-5 mg of protein lysate with 50 μL OtUBD resin for 2 hours at 4°C with end-over-end mixing.
- Wash resin sequentially with:
- 10 column volumes of Lysis Buffer
- 10 column volumes of High-Salt Buffer (50 mM Tris-HCl pH 7.5, 500 mM NaCl, 0.1% NP-40)
- 10 column volumes of No-Salt Buffer (50 mM Tris-HCl pH 7.5)
- Elubiquitinated proteins with 2-3 column volumes of Elution Buffer at 65°C for 10 minutes.
Downstream Applications:
- For immunoblotting: Separate eluates by SDS-PAGE and probe with anti-ubiquitin antibodies.
- For proteomics: Process eluates for LC-MS/MS analysis, including digestion, peptide cleanup, and mass spectrometry.
This protocol has been successfully applied to both baker's yeast and mammalian cell lysates, demonstrating its broad utility across model systems [24]. The inclusion of NEM at 20 mM is critical for preserving ubiquitination by inhibiting deubiquitinating enzymes, a common challenge when working with low-stoichiometry ubiquitination events.
TUBE-MS for Monitoring Small Molecule-Induced Ubiquitination Changes
The TUBE-MS method enables quantitative assessment of changes in protein polyubiquitination in response to small molecule treatments, such as PROTACs or DUB inhibitors [35].
Materials and Reagents:
- Biotinylated TUBE (tandem ubiquitin-binding entities)
- Magnetic streptavidin beads
- Lysis Buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 1 mM EDTA, 4 M urea, 20 mM NEM
- Wash Buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% NP-40, 4 M urea
- Elution Buffer: 50 mM Tris-HCl (pH 7.5), 2% SDS, 5% β-mercaptoethanol
Step-by-Step Protocol:
Cell Treatment and Lysis:
- Treat cells with compound of interest (e.g., PROTAC, DUB inhibitor) for appropriate duration.
- Optionally pre-incubate with proteasome inhibitor (e.g., Carfilzomib) for 1 hour to stabilize ubiquitinated proteins.
- Lyse cells in urea-containing Lysis Buffer with 20 mM NEM to inhibit DUBs and dissociate non-covalent interactions.
Polyubiquitin Enrichment:
- Incubate 2-5 mg clarified lysate with biotinylated TUBE (10-20 μg) for 2 hours at 4°C.
- Add magnetic streptavidin beads and incubate for 1 hour.
- Wash beads sequentially with:
- 10 volumes of Wash Buffer
- 10 volumes of No-Detergent Buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl)
- Elute bound proteins with Elution Buffer at 65°C for 10 minutes.
LC-MS/MS Analysis:
- Digest eluted proteins with trypsin.
- Clean up peptides using C18 stage tips.
- Analyze by LC-MS/MS using data-dependent acquisition.
- Process data using standard proteomics software (MaxQuant, Proteome Discoverer).
This TUBE-MS workflow has been successfully applied to identify ubiquitination changes induced by diverse compounds, including the PROTAC MZ1 and USP7 inhibitors, revealing both degradative and non-degradative ubiquitination events [35]. The semi-denaturing conditions with 4 M urea are essential for reducing co-enrichment of non-ubiquitinated proteins and Ub-binding partners.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagent Solutions for UBD-Based Enrichment
| Reagent | Function | Application Notes |
|---|---|---|
| N-Ethylmaleimide (NEM) [35] [34] | Irreversible cysteine protease/DUB inhibitor | Critical for preserving ubiquitination; use at 20 mM during lysis |
| cOmplete EDTA-free Protease Inhibitor Cocktail [24] | Inhibits serine, threonine, and metalloproteases | Prevents general protein degradation during enrichment |
| SulfoLink Coupling Resin [24] | Support for covalent OtUBD immobilization | Stable affinity matrix with high binding capacity |
| Magnetic Streptavidin Beads [35] | Solid support for biotinylated TUBEs | Enable rapid processing and automation compatibility |
| Linkage-Specific Ub Antibodies [19] | Detection of specific ubiquitin chain types | Validate chain linkage specificity after enrichment |
Workflow Visualization: Strategic Approaches to Enrichment
The following diagrams illustrate key experimental workflows and technology comparisons for UBD-based enrichment of low-stoichiometry ubiquitination events.
Diagram 1: Generalized workflow for UBD-based enrichment of ubiquitinated proteins, highlighting critical steps for combating low stoichiometry, including DUB inhibition during sample preparation and stringent wash conditions.
Diagram 2: Technology selection guide matching UBD-based enrichment approaches to specific research objectives, highlighting how each technology addresses different aspects of the low-stoichiometry challenge.
UBD-based affinity enrichment methods have dramatically advanced our ability to study low-stoichiometry ubiquitination events, which are crucial for understanding ubiquitin signaling in health and disease. The development of high-affinity tools like OtUBD, TUBEs, and ThUBD has enabled researchers to overcome the fundamental challenge of low abundance, providing the sensitivity and specificity required for comprehensive ubiquitinome analysis.
As the field progresses, several emerging trends promise to further enhance our capabilities. The integration of UBD-based enrichment with advanced mass spectrometry techniques, including data-independent acquisition and targeted proteomics, will improve the quantification of ubiquitination dynamics. Additionally, the growing emphasis on high-throughput methodologies reflects the increasing importance of ubiquitination profiling in drug discovery, particularly for characterizing PROTACs and other targeted protein degradation therapeutics [1] [36]. Finally, the continued development of linkage-specific UBDs will enable more precise decoding of the ubiquitin code, revealing how specific chain architectures regulate cellular physiology.
By implementing the detailed protocols and strategic approaches outlined in this application note, researchers can effectively combat the challenges of low-stoichiometry ubiquitination, advancing both basic science and therapeutic development in this critical field.
The ubiquitin-proteasome system (UPS) is a crucial regulatory mechanism for maintaining cellular protein homeostasis, governing the degradation of proteins to influence nearly all cellular processes [37]. A critical and often targeted component of this system is the deubiquitinating enzyme (DUB), a class of proteases responsible for cleaving ubiquitin from substrate proteins, thereby reversing the signal for degradation [38]. The dynamic balance between ubiquitination by E3 ligases and deubiquitination by DUBs allows for precise control over protein stability and function [37]. Inhibiting DUB activity is essential in experimental settings to preserve ubiquitin signals on substrate proteins, enabling researchers to study these modifications and their functional consequences. This application note details the use of DUB inhibitors, with a specific focus on the broad-spectrum cysteine protease inhibitor N-Ethylmaleimide (NEM), within the context of affinity enrichment methods for studying ubiquitination.
The Mechanism of N-Ethylmaleimide as a DUB Inhibitor
N-Ethylmaleimide (NEM) is an organic compound that functions as an irreversible inhibitor of cysteine peptidases, a class that includes the majority of DUBs [39] [40]. Its mechanism of action is characterized by its role as a Michael acceptor, which allows it to rapidly and covalently alkylate the thiol group (-SH) of cysteine residues within the enzyme's active site [40]. This alkylation reaction forms a stable thioether bond that is virtually irreversible under standard experimental conditions, leading to the permanent inactivation of the enzyme [39] [40]. This property makes NEM exceptionally effective for the complete inactivation of endogenous DUBs during cell lysis and protein extraction, preventing the undesired loss of ubiquitin chains from proteins of interest before they can be isolated and analyzed [41]. NEM is widely used in lysis buffers, typically at concentrations around 5-20 mM, to ensure robust inhibition of deubiquitination and de-sumoylation activities, thereby preserving the native ubiquitination state of proteins for downstream applications like western blotting or mass spectrometry [41] [40].
Table 1: Key Characteristics of N-Ethylmaleimide (NEM)
| Property | Description |
|---|---|
| Chemical Name | 1-Ethyl-1H-pyrrole-2,5-dione |
| Molecular Weight | 125.13 g/mol |
| Mechanism | Irreversible cysteine protease inhibitor; alkylates active site thiol group [39] |
| Primary Use in UPS | Inactivates endogenous deubiquitinating enzymes (DUBs) during sample preparation [39] [41] |
| Reaction Specificity | Reacts with thiols at pH 6.5–7.5; can react with amines or hydrolyze at alkaline pH [40] |
| Common Working Concentration | 5-20 mM in lysis buffers [41] |
Experimental Protocol for Ubiquitinated Protein Enrichment with NEM
The following protocol is adapted from established methods for the affinity purification of ubiquitinated proteins from mammalian cells, highlighting the critical steps where NEM is required to prevent deubiquitination [41].
Primary Instruments and Equipment
- High-speed centrifuge
- Ultracentrifuge
- Ultrasonic crusher or probe sonicator
- Vertical shaker
- Metal bath or water bath
Reagents and Solutions
- Guanidine Hydrochloride Lysis Solution: 6 M Guanidine hydrochloride, 100 mM Sodium phosphate buffer (pH 8.0), 5 mM Imidazole.
- Guanidine Hydrochloride Wash Buffer with NEM: 6 M Guanidine hydrochloride, 50 mM Sodium phosphate buffer (pH 8.0), 10 mM Tris-Cl (pH 8.0), 300 mM NaCl, 5 mM N-Ethylmaleimide (NEM). Note: NEM should be added fresh before use. [41]
- Ni²⁺-NTA-agarose beads
- Protein Buffer: 50 mM Sodium phosphate buffer (pH 8.0), 100 mM KCl, 20% (v/v) Glycerol, 0.2% (v/v) NP-40.
- Elution Buffer: Protein buffer containing 200 mM imidazole.
- Protease Inhibitor Cocktail (e.g., PMSF, EDTA, Pepstatin, Leupeptin) [41].
Step-by-Step Procedure
- Cell Lysis: Harvest cultured mammalian cells expressing His₆-tagged ubiquitin (His₆-Ub) and a protein of interest. Lyse the cells in Guanidine Hydrochloride Lysis Solution. To reduce viscosity, perform brief sonication using a probe sonicator [41].
- Clarification: Centrifuge the lysate at 14,000 × g for 15 minutes at 4°C. Transfer the clarified supernatant to a new tube.
- Affinity Binding: Incubate the clarified lysate with a pre-determined volume of Ni²⁺-NTA-agarose beads (e.g., 75 µL) for 4 hours at 4°C on a vertical shaker.
- Washing: Transfer the bead-lysate mixture to a disposable column. Wash the beads sequentially with the following buffers to remove non-specifically bound proteins [41]:
- 1 mL of 6 M guanidine hydrochloride/100 mM sodium phosphate buffer (pH 8.0), without imidazole.
- 2 mL of 6 M guanidine hydrochloride/100 mM sodium phosphate buffer (pH 5.8).
- 1 mL of 6 M guanidine hydrochloride/100 mM sodium phosphate buffer (pH 8.0), without imidazole.
- 2 mL of a 1:1 (v/v) mixture of 6 M guanidine hydrochloride/100 mM sodium phosphate buffer (pH 8.0) and protein buffer without imidazole.
- 2 mL of a 1:3 (v/v) mixture of 6 M guanidine hydrochloride/100 mM sodium phosphate buffer (pH 8.0) and protein buffer without imidazole.
- 2 mL of protein buffer without imidazole.
- 1 mL of protein buffer containing 10 mM imidazole.
- Elution: Elute the bound ubiquitinated proteins with 1 mL of Elution Buffer (protein buffer containing 200 mM imidazole).
- Precipitation and Analysis: Precipitate the eluate with 10% (v/v) trichloroacetic acid (TCA). Resuspend the precipitate in 2× SDS-PAGE loading buffer and denature in a boiling water bath for 5 minutes. The samples can now be analyzed by SDS-PAGE and western blotting, or prepared for mass spectrometry to identify ubiquitination sites [41].
Diagram 1: Ubiquitinated Protein Enrichment Workflow. Steps critical for DUB inhibition with NEM are highlighted in red.
Quantitative Data on DUB Inhibitors
While NEM is a classic, broad-spectrum inhibitor, the field of DUB research has expanded to include more selective compounds. The table below summarizes key inhibitors, illustrating the progression from non-selective tools to targeted therapeutic candidates.
Table 2: Profile of Selected Deubiquitinase (DUB) Inhibitors
| Inhibitor Name | Primary Target(s) | Mechanism / Key Characteristic | Reported IC₅₀ / Activity | Therapeutic Application Focus |
|---|---|---|---|---|
| N-Ethylmaleimide (NEM) | All cysteine peptidases / Broad-spectrum DUBs [39] | Irreversible alkylation of active site cysteine [40] | Inactivates at 5-20 mM in buffers [41] | Research tool for sample preparation |
| P022077 | USP7 (Ubiquitin-Specific Protease 7) [38] | Selective small-molecule inhibitor | Characterized via multiplex assay [38] | Oncology (discovery stage) |
| PR-619 | Multiple DUBs (Broad-spectrum) [38] | Cell-permeable, reversible inhibitor | Used in vitro for characterization [38] | Chemical biology tool compound |
| KSQ-4279 | USP1 (Ubiquitin-Specific Protease 1) [42] | Clinical-stage small-molecule inhibitor | Preclinical and clinical studies [42] | Oncology (e.g., combination therapies) |
The Scientist's Toolkit: Key Research Reagent Solutions
Successful research into ubiquitination requires a suite of reliable reagents. The following table outlines essential tools for experiments involving the prevention of deubiquitination.
Table 3: Essential Research Reagents for Ubiquitination Studies
| Reagent / Material | Function and Importance in Ubiquitination Research |
|---|---|
| N-Ethylmaleimide (NEM) | Irreversibly inhibits cysteine-based DUBs during cell lysis and protein extraction, preserving the ubiquitinated state of proteins for analysis [39] [41]. |
| His₆-Tagged Ubiquitin | Allows for high-affinity purification of ubiquitinated proteins from cell lysates using nickel-based (Ni²⁺-NTA) chromatography, a cornerstone of enrichment protocols [41]. |
| Ni²⁺-NTA-Agarose Beads | The solid-phase affinity resin for binding and purifying polyhistidine-tagged proteins (e.g., His₆-Ub conjugates) from complex lysates [41]. |
| Protease Inhibitor Cocktails | Broad-spectrum mixtures (e.g., containing PMSF, EDTA) that inhibit serine, aspartic, and metalloproteases, working alongside NEM to ensure overall protein integrity [41]. |
| Ubiquitin Binding Domains (UBDs) | Modular protein domains (e.g., UBA, UIM, NZF) used as reagents to detect, affinity-purify, or characterize specific ubiquitin chain types and linkages [7] [6]. |
| Selective DUB Inhibitors | Small molecules (e.g., P022077 for USP7) that target specific DUB families, enabling functional studies of individual DUBs in cells without broad DUB shutdown [38] [42]. |
The use of DUB inhibitors, from broad-spectrum tools like N-Ethylmaleimide to newly developed selective compounds, is indispensable for dissecting the complex roles of the ubiquitin-proteasome system. The rigorous application of NEM in sample preparation protocols ensures the integrity of ubiquitin signals, forming the foundation for accurate analysis via UBD-based affinity enrichment methods. As research progresses, the integration of these classical biochemical tools with modern, high-selectivity inhibitors will continue to drive discoveries in both basic biology and the development of targeted therapies for diseases such as cancer and neurodegenerative disorders [43] [42] [37].
Diagram 2: DUB Inhibitors Maintain Ubiquitin Signaling. DUBs reverse ubiquitination; their inhibition preserves the signal for degradation or other functions.
In the study of ubiquitin signaling, affinity enrichment methods using ubiquitin-binding domains (UBDs) are powerful tools for mapping the ubiquitinome. However, the success of these techniques is critically dependent on the initial cell lysis conditions. The core challenge lies in optimizing lysis buffers to achieve efficient disruption of cellular membranes while simultaneously preserving the labile, non-covalent interactions between ubiquitinated proteins and their binding partners. This balance is essential for accurate downstream analysis, whether the goal is to profile the covalent ubiquitinome or to capture the broader ubiquitin interactome. This application note provides a detailed, practical framework for selecting and optimizing lysis buffers specifically for UBD-based affinity enrichment protocols, complete with quantitative data and validated experimental procedures.
The Critical Role of Lysis Conditions in Ubiquitin Enrichment
Ubiquitination is a dynamic and reversible post-translational modification that regulates nearly all cellular processes, from protein degradation to immune responses and DNA repair [1]. The ubiquitin-proteasome system (UPS) serves as the core machinery for targeted protein degradation in eukaryotes, and its dysregulation is linked to major human diseases, including cancers and neurodegenerative disorders [1]. UBD-based affinity enrichment, using tools such as the high-affinity OtUBD or engineered tandem hybrid UBDs (ThUBDs), has emerged as a versatile method for capturing ubiquitinated proteins from complex biological samples [4] [44]. These methods can be performed under native (non-denaturing) or denaturing conditions, with the choice dictating whether non-covalently associated proteins are co-purified.
The lysis buffer is the first and one of the most determinative variables in this workflow. An overly harsh, denaturing buffer will efficiently solubilize cellular contents, including membrane proteins, but will disrupt non-covalent protein-protein interactions, providing a snapshot of only covalently ubiquitinated proteins. Conversely, a mild, non-denaturing buffer preserves the native ubiquitin interactome but may suffer from lower lysis efficiency and fail to solubilize all cellular compartments. The composition of the buffer—specifically the type and concentration of detergents and chaotropic agents—directly influences this balance, impacting the yield, specificity, and ultimately, the biological relevance of the enrichment results [45] [46].
Comparative Analysis of Lysis Buffer Components
To make an informed choice on lysis conditions, it is essential to understand the properties and trade-offs of common buffer components. The table below summarizes key reagents and their effects on lysis efficiency and interaction preservation.
Table 1: Key Components of Cell Lysis Buffers and Their Applications
| Component | Type | Mechanism of Action | Advantages | Disadvantages | Ideal for UBD-based Enrichment of: |
|---|---|---|---|---|---|
| SDS | Ionic Detergent | Disrupts lipid bilayers; denatures proteins [45]. | High efficiency for membrane protein solubilization [45]. | Difficult to remove; interferes with MS and enzyme activity; disrupts all non-covalent interactions [45]. | Covalent Ubiquitinome (Denaturing Workflows) |
| Guanidinium HCl (GnHCl) | Chaotrope | Denatures proteins by disrupting hydrogen bonds [45]. | Compatible with LC-MS analysis; does not precipitate like SDS [45]. | Denatures proteins, disrupting interactions. | Covalent Ubiquitinome (Denaturing Workflows) |
| NP-40/Triton X-100 | Non-ionic Detergent | Solubilizes membrane lipids and proteins [46]. | Mild, non-denaturing; preserves protein interactions and activity [46]. | Lower efficiency for some membrane proteins. | Ubiquitin Interactome (Native Workflows) |
| Sodium Deoxycholate | Ionic Detergent | Solubilizes and denatures proteins [46]. | Effective for total protein extraction. | Can disrupt protein interactions; may interfere with some downstream assays. | Covalent Ubiquitinome (Moderately Denaturing) |
| RIPA Buffer | Mixed Detergent Buffer | Contains both ionic (e.g., deoxycholate) and non-ionic (e.g., NP-40) detergents, sometimes with SDS [46]. | Effective for extracting nuclear and membrane proteins. | Can be partially denaturing, potentially disrupting some weak interactions. | Context-Dependent |
The choice of lysis buffer directly impacts proteome coverage. A comparative study on HeLa cells and human plasma found that the combination of a powerful detergent like SDS with the SP3 (single-pot, solid-phase-enhanced sample preparation) digestion method resulted in the highest number of quantified proteins (6131 ± 20 proteins), outperforming GnHCl-based in-solution digestion (ISD), which identified 4851 ± 44 proteins [45]. Crucially, the SP3/SDS workflow was particularly effective at quantifying membrane-associated proteins, which are often involved in critical signaling pathways [45]. This demonstrates that for a comprehensive view of the covalent ubiquitinome, including challenging membrane proteins, a stronger denaturant is beneficial.
Table 2: Quantitative Performance of Different Lysis and Digestion Workflows in HeLa Cells
| Workflow | Lysis Buffer | Number of Quantified Proteins (Mean ± SEM) | Number of Quantified Peptides (Mean ± SEM) | Peptides with Zero Missed Cleavages (%) |
|---|---|---|---|---|
| SP3 | SDS | 6131 ± 20 | 47,088 ± 345 | 84.6% |
| SP3 | Guanidinium HCl | 5895 ± 37 | 48,940 ± 345 | 77.5% |
| In-Solution Digestion (ISD) | Guanidinium HCl | 4851 ± 44 | 40,505 ± 630 | 38.0% |
Optimized Protocols for UBD-Based Enrichment
The following protocols are adapted from established methods for OtUBD and ThUBD, providing two pathways tailored for different research objectives [4] [44].
Protocol A: Native Lysis for Enriching the Ubiquitin Interactome
This protocol is designed to isolate ubiquitinated proteins along with their non-covalently bound interaction partners.
- Step 1: Cell Lysis
- Lysis Buffer Formulation: 50 mM Na₂HPO₄ (pH 8.0), 500 mM NaCl, 0.01% SDS, 5% glycerol [44]. Note: The SDS concentration here is very low and is used to aid solubilization without fully denaturing complexes.
- Procedure: Harvest and wash cells. Resuspend the cell pellet in ice-cold lysis buffer supplemented with protease and deubiquitinase (DUB) inhibitors. Incubate on ice for 15-30 minutes with occasional vortexing. For tissue samples, mechanical homogenization in a buffer like T-PER is recommended [46].
- Step 2: Clarification
- Centrifuge the lysate at high speed (e.g., 70,000 × g for 30 minutes at 4°C) to remove insoluble debris [44]. Transfer the clear supernatant to a new tube.
- Step 3: Affinity Enrichment
- Step 4: Washing
- Pellet the beads and wash sequentially with lysis buffer and a mild wash buffer (e.g., 50 mM NH₄HCO₃) to remove non-specifically bound proteins [44].
- Step 5: Elution
- Elute the bound ubiquitinated proteins and their complexes by boiling the beads in 1× SDS-PAGE loading buffer or by competitive elution with free ubiquitin for downstream analysis [44].
Protocol B: Denaturing Lysis for Specific Enrichment of the Covalent Ubiquitinome
This protocol uses strong denaturants to isolate only proteins that are covalently modified by ubiquitin.
- Step 1: Cell Lysis
- Step 2: Clean-up and Digestion Compatibility (for SDS samples)
- SDS must be removed prior to mass spectrometry. Use methods like SP3, which involves transferring the lysate to a fresh tube and adding paramagnetic beads in high ethanol concentration (e.g., 80%) to bind proteins and wash away SDS [45].
- Step 3: Affinity Enrichment
- Dilute the denatured lysate (or the cleaned-up protein sample) into a compatible binding buffer. Proceed with incubation on UBD-conjugated beads as in Protocol A [4].
- Step 4: Washing and Elution
- Wash and elute as described in Protocol A. The use of a denaturing lysis buffer ensures that proteins eluted are primarily those directly bound to the UBD via ubiquitin, not indirect interactors.
The workflow for selecting and applying these protocols is summarized in the diagram below.
The Scientist's Toolkit: Essential Research Reagents
Successful execution of these protocols requires a set of defined reagents and tools. The following table lists key solutions and materials for UBD-based ubiquitin enrichment studies.
Table 3: Research Reagent Solutions for UBD-based Affinity Enrichment
| Reagent / Material | Function / Application | Examples / Key Characteristics |
|---|---|---|
| High-Affinity UBD Resin | Core affinity matrix for capturing ubiquitinated proteins. | OtUBD affinity resin [4]; Engineered Tandem Hybrid UBDs (ThUBDs) like ThUDA20 with high, unbiased affinity for ubiquitin chains [44] [1]. |
| Native Lysis Buffer | Cell lysis while preserving protein interactions. | NP-40 Lysis Buffer [46]; Modified RIPA (without SDS) for immunoprecipitation [46]; 50 mM Na₂HPO₄, 500 mM NaCl, 0.01% SDS, 5% glycerol [44]. |
| Denaturing Lysis Buffer | Complete solubilization and inactivation of enzymes. | RIPA Lysis Buffer (contains ionic detergents) [46]; Buffers with 1-4% SDS or 4-6 M Guanidinium HCl [4] [45]. |
| Protease & DUB Inhibitors | Prevent protein degradation and deubiquitination during lysis. | Halt Protease Inhibitor Cocktail; specific DUB inhibitors. Essential for all workflows to maintain ubiquitin modifications [46]. |
| SP3 Paramagnetic Beads | Efficient protein clean-up and digestion, especially for SDS-lysed samples. | Enable SDS removal and high-efficiency tryptic digestion, leading to superior proteome coverage [45]. |
| Depletion Spin Columns | Increase proteome coverage in complex samples like plasma. | Remove highly abundant proteins (e.g., albumin), allowing quantification of lower-abundance proteins [45]. |
Buffer optimization is not a one-size-fits-all process but a strategic decision that directly shapes research outcomes in ubiquitin biology. By selecting a native lysis buffer, researchers can capture the dynamic network of the ubiquitin interactome. In contrast, a denaturing approach provides a focused, high-efficiency view of the covalent ubiquitinome, which is particularly powerful for discovering direct ubiquitination targets and mapping modification sites via mass spectrometry. The protocols and data presented here provide a clear roadmap for researchers to tailor their lysis conditions, thereby ensuring that their UBD-based affinity enrichment strategies yield biologically meaningful and technically robust results.
In the study of ubiquitin biology, affinity enrichment methods utilizing ubiquitin-binding domains (UBDs) are indispensable for deciphering the ubiquitinome and ubiquitin interactome. However, the efficacy of these techniques is critically dependent on minimizing non-specific interactions, which can obscure true biological signals and lead to erroneous conclusions. Non-specific binding compromises sample purity, reduces the yield of target ubiquitinated proteins, and impedes downstream analyses such as immunoblotting and mass spectrometry. The strategic selection of chromatography resins and the meticulous formulation of wash buffers are therefore foundational to successful experimental outcomes. This application note provides a detailed framework for optimizing these parameters, with a specific focus on UBD-based affinity enrichment, to achieve high-purity ubiquitinome profiling.
Background: UBD-Based Affinity Enrichment
Ubiquitin-binding domains are protein modules that recognize and non-covalently interact with ubiquitin or ubiquitin chains. Their application in affinity purification has revolutionized the study of protein ubiquitination. Unlike antibody-based methods, which can be expensive and exhibit sequence bias, UBD-based approaches, particularly those using high-affinity domains like OtUBD from Orientia tsutsugamushi, offer a versatile and economical alternative [24] [4]. The OtUBD domain exhibits low nanomolar affinity for ubiquitin and can enrich both mono- and poly-ubiquitinated proteins from complex lysates, a significant advantage over methods like Tandem Ubiquitin-Binding Entities (TUBEs), which are less effective for monoubiquitinated species [24] [47].
A critical consideration in any ubiquitin enrichment protocol is the preservation of the native ubiquitination state. Deubiquitylases (DUBs) are highly active and can rapidly reverse ubiquitination during cell lysis and purification. Therefore, the use of potent DUB inhibitors in all buffers is mandatory. Research indicates that while concentrations of 5-10 mM of alkylating agents like N-ethylmaleimide (NEM) or iodoacetamide (IAA) are common, up to 10-fold higher concentrations may be necessary to fully preserve certain ubiquitin chains, such as K63- and M1-linked polymers [48]. Furthermore, for experiments involving mass spectrometry, NEM is preferred over IAA because its adduct does not interfere with the detection of the Gly-Gly remnant left on ubiquitinated lysines after tryptic digestion [48].
Resin Selection for UBD-Based Purification
The solid support, or resin, to which the UBD is immobilized, plays a pivotal role in determining the binding capacity, specificity, and physical robustness of the purification process.
Table 1: Key Properties of Common Affinity Chromatography Resins
| Resin Type | Material | Bead Size | Key Advantages | Considerations for Ubiquitin Enrichment |
|---|---|---|---|---|
| Crosslinked Beaded Agarose [49] | 4% or 6% Agarose | 45-165 µm | Low non-specific binding; excellent for gravity-flow and low-pressure applications. | The standard choice for most applications; may compress under higher pressure. |
| Superflow Agarose [49] | Highly crosslinked Agarose | 45-165 µm | Improved flow rates and pressure tolerance; maintains high binding capacity. | Suitable for methods requiring faster processing or slight pressure. |
| UltraLink Biosupport [49] | Polyacrylamide | Not specified in results | High mechanical stability; low non-specific binding; resistant to compression. | Ideal for medium-pressure systems and when exceptional durability is needed. |
| SulfoLink Coupling Resin [24] | Crosslinked beaded agarose | Not specified in results | Specifically designed for covalent coupling via sulfhydryl groups. | Used in the OtUBD protocol for immobilizing cysteine-containing constructs [24]. |
For most UBD-based purifications, crosslinked beaded agarose resins like 4% or 6% agarose CL-4B/CL-6B are the default and highly effective choice due to their high porosity and minimal non-specific binding [49]. The selection of a specific resin should be guided by the scale of the experiment and the required flow characteristics.
Wash Buffer Optimization Strategies
The wash step is the primary opportunity to remove non-specifically bound contaminants while leaving the target ubiquitinated proteins bound to the UBD resin. A strategic, multi-stage wash protocol is recommended.
Table 2: Wash Buffer Formulations for Minimizing Non-Specific Binding
| Wash Buffer Type | Example Formulation | Primary Mechanism of Action | Application Notes |
|---|---|---|---|
| Low-Detergent Wash [24] [49] | PBS or Tris buffer with 0.1% to 0.5% Triton X-100 or Tween-20 | Disrupts hydrophobic and ionic interactions without denaturing proteins. | Initial wash under native conditions to remove weakly associated proteins from the ubiquitin interactome. |
| Moderate Salt Wash [24] [50] | 20-300 mM NaCl in standard binding buffer (e.g., PBS) | Disrupts ionic interactions by shielding charges on proteins and the resin. | Effective for reducing host cell protein (HCP) contamination without eluting the target [50]. |
| Chaotrope Wash (Denaturing) [24] [41] | 8 M Urea, 50 mM sodium phosphate (pH 8.0), 300 mM NaCl | Denatures and solubilizes proteins, effectively disrupting strong non-covalent interactions. | Used in denaturing workflows to specifically isolate covalently ubiquitinated proteins from non-covalent interactors [24]. |
| High-Stringency Wash [41] | 6 M Guanidine hydrochloride, 50 mM sodium phosphate (pH 8.0) | Powerful denaturant that effectively strips nearly all non-covalently bound proteins. | A very harsh wash for the most challenging contamination; may require subsequent resin re-equilibration. |
Executing a Tiered Wash Protocol
- Primary Wash: Begin with 3-5 column volumes of a physiological buffer like PBS containing 0.1% detergent (e.g., Triton X-100) to remove the bulk of soluble contaminants.
- Secondary Wash: Follow with 3-5 column volumes of a moderate salt buffer (e.g., PBS with 150-300 mM NaCl) to disrupt electrostatic non-specific binding.
- Tertiary Wash (if applicable): For denaturing protocols aimed at isolating the direct ubiquitinome, perform 3-5 column volumes of a chaotrope-containing wash buffer (e.g., with 8 M Urea) [24] [41]. Always include DUB inhibitors like 5-50 mM NEM in all wash buffers to prevent deubiquitination [24] [48].
Detailed Experimental Protocol: OtUBD-Based Enrichment
The following protocol, adapted from established methods, details the enrichment of ubiquitinated proteins from mammalian cell lysates using OtUBD affinity resin under both native and denaturing conditions [24].
Materials and Reagents
Table 3: Research Reagent Solutions for UBD-Based Enrichment
| Reagent / Kit | Function / Application | Example Source / Catalog Number |
|---|---|---|
| pET21a-cys-His6-OtUBD Plasmid [24] | Recombinant production of cysteine- and histidine-tagged OtUBD. | Addgene, plasmid #190091 |
| SulfoLink Coupling Resin [24] | Immobilizes the purified OtUBD via its cysteine residue to create the affinity resin. | Thermo Scientific, catalog #20402 |
| cOmplete EDTA-free Protease Inhibitor Cocktail [24] | Inhibits proteolytic degradation of samples. | Roche, catalog #11873580001 |
| N-Ethylmaleimide (NEM) [24] [48] | Alkylating agent; irreversibly inhibits deubiquitylases (DUBs) to preserve ubiquitination. | Sigma-Aldrich, catalog #E3876 |
| Tris(2-carboxyethyl)phosphine (TCEP) [24] | Reducing agent; maintains cysteine residues in a reduced state for coupling. | Sigma-Aldrich, catalog #C4706 |
| Polyubiquitin Affinity Resin [41] | Alternative resin for ubiquitin enrichment; can be used for comparison. | Pierce Inc. |
Step-by-Step Procedure
Part A: Preparation of OtUBD Affinity Resin
- Express and Purify OtUBD: Transform E. coli with the pET21a-cys-His6-OtUBD plasmid. Induce expression with IPTG. Purify the recombinant protein using Ni-NTA agarose chromatography under standard native conditions [24].
- Immobilize OtUBD: Reduce the purified OtUBD with 1 mM TCEP. Incubate the reduced protein with SulfoLink coupling resin according to the manufacturer's instructions. Block any remaining reactive groups with L-cysteine.
- Store the Resin: Store the prepared OtUBD resin in PBS with 0.02% sodium azide at 4°C.
Part B: Cell Lysis and Affinity Pulldown
- Harvest and Lyse Cells: Culture and harvest mammalian cells (e.g., HEK293T or HeLa). Lyse cells in an appropriate volume of Lysis Buffer.
- Clarify Lysate: Centrifuge the lysate at >14,000 × g for 15 minutes at 4°C. Collect the supernatant.
- Incubate Lysate with Resin: Transfer a measured volume of clarified lysate (e.g., 1-2 mg total protein) to a tube containing a pre-determined amount of OtUBD resin (e.g., 20-50 µL bed volume). Incubate on a rotator or vertical shaker for 2 hours to overnight at 4°C [24] [41].
- Wash the Resin: Pellet the resin by brief centrifugation and carefully remove the supernatant. Perform the tiered wash protocol: a. Wash 3 times with 10-20 resin volumes of Wash Buffer I (e.g., PBS with 0.1% Triton X-100). b. Wash 2 times with 10-20 resin volumes of Wash Buffer II (e.g., PBS with 0.1% Triton X-100 and 300 mM NaCl). c. (For denaturing protocol only) Wash 2 times with 10-20 resin volumes of Wash Buffer III (e.g., 8 M Urea, 50 mM sodium phosphate pH 8.0, 300 mM NaCl) [24] [41].
- Elute Ubiquitinated Proteins: After the final wash, elute the bound proteins by incubating the resin with 2-3 resin volumes of SDS-PAGE loading buffer containing 50-100 mM DTT at 95°C for 5-10 minutes. Alternatively, for gentle elution, competitive elution with free ubiquitin (0.5-1 mg/mL) can be used [24].
Workflow and Pathway Diagrams
The following diagram illustrates the key decision points and steps in the optimized protocol for enriching ubiquitinated proteins.
Diagram 1: Experimental workflow for ubiquitinated protein enrichment, highlighting critical steps for minimizing non-specific binding.
The ubiquitin signaling pathway involves a cascade of enzymes that lead to diverse cellular outcomes, which UBD-based methods aim to decipher.
Diagram 2: The ubiquitin signaling cascade and the role of UBD-based enrichment in detecting its products.
The strategic optimization of resin selection and wash buffer formulations is not merely a technical exercise but a critical component of robust ubiquitin research. The implementation of a tiered wash strategy, progressing from low to high stringency, systematically displaces contaminants based on the strength of their non-specific interactions. Coupled with the choice of a high-quality, low-binding resin like beaded agarose, this approach significantly enhances the signal-to-noise ratio in downstream analyses.
As evidenced by the successful application of the OtUBD protocol, these principles enable researchers to distinguish between the covalently modified ubiquitinome and the non-covalent ubiquitin interactome with high confidence [24]. The recommendations provided here, including the use of high concentrations of DUB inhibitors and tailored buffer formulations, serve as a foundation for generating reliable, high-quality data. Adherence to these optimized methods ensures that researchers can accurately capture the complex landscape of protein ubiquitination, thereby advancing our understanding of its vital role in cellular regulation and disease.
Ubiquitination is an essential post-translational modification that regulates diverse cellular functions, including protein degradation, DNA repair, and cell signaling [19]. The versatility of ubiquitin signaling arises from the complexity of ubiquitin conjugates, which range from single ubiquitin monomers to polymers with different lengths and linkage types [19]. Defective ubiquitination has been associated with many diseases, including neurodegenerative disorders and various cancers, making the identification of ubiquitinated proteins a key step in understanding cellular regulatory mechanisms and developing targeted therapies [24] [19].
Enrichment of ubiquitinated proteins from complex biological samples presents significant challenges due to the low stoichiometry of modification, the diversity of ubiquitin chain architectures, and the transient nature of ubiquitin-protein interactions [19]. Among the various enrichment strategies, ubiquitin-binding domain (UBD)-based affinity purification has emerged as a powerful tool that offers advantages over traditional antibody-based approaches or tagged ubiquitin expression systems [24] [19]. This application note provides a comprehensive decision framework for selecting between native and denaturing workflows in UBD-based ubiquitin enrichment, supported by experimental data and detailed protocols.
Table 1: Key Comparison of Ubiquitin Enrichment Methodologies
| Method | Principles | Advantages | Limitations |
|---|---|---|---|
| Epitope-Tagged Ubiquitin | Expression of affinity-tagged Ub; purification using anti-epitope antibodies | Relatively low-cost; easy implementation | May cause spurious ubiquitination patterns; not feasible for clinical tissues [19] |
| Ub Antibody-Based | Antibodies recognizing ubiquitin or specific linkages enrich conjugates | Works with endogenous ubiquitin; applicable to tissues | High cost; potential non-specific binding [24] [19] |
| UBD-Based Affinity | Ubiquitin-binding domains capture ubiquitinated proteins | Versatile and economical; works with all ubiquitin conjugates | Requires optimization of binding conditions [24] |
Technical Comparison: Native Versus Denaturing Workflows
Fundamental Principles and Applications
The choice between native and denaturing workflows fundamentally depends on the research objectives—specifically, whether the goal is to capture the complete ubiquitin interactome (including non-covalent interacting partners) or to isolate specifically the covalently modified ubiquitinome [24].
Native conditions preserve the non-covalent interactions between ubiquitin/ubiquitinated proteins and ubiquitin-binding proteins, enabling the study of protein complexes and signaling assemblies [24]. This approach is particularly valuable for investigating the functional consequences of ubiquitination in cellular pathways. In contrast, denaturing conditions disrupt non-covalent interactions while preserving the isopeptide bonds of ubiquitin modifications, allowing researchers to focus specifically on directly ubiquitinated proteins without co-purifying interactors [24]. This distinction is crucial for accurate ubiquitin site mapping and understanding direct versus indirect ubiquitination events.
Performance Characteristics and Limitations
Recent methodological advances have revealed significant differences in performance between these approaches. The Denatured-Refolded Ubiquitinated Sample Preparation (DRUSP) method, which incorporates a denaturing and refolding step before UBD-based enrichment, demonstrates a significantly stronger ubiquitin signal—nearly three times greater than conventional native methods [5]. When combined with tandem hybrid UBDs, DRUSP improved overall ubiquitin signal enrichment by approximately 10-fold compared to control methods [5].
Table 2: Quantitative Performance Comparison of Enrichment Workflows
| Parameter | Native Workflow | Conventional Denaturing | DRUSP with Tandem UBD |
|---|---|---|---|
| Ubiquitin Signal Intensity | Baseline | ~3x baseline [5] | ~10x baseline [5] |
| Identification of Mono-Ubiquitination | Effective with high-affinity UBDs [24] | Limited | Enhanced after refolding |
| Identification of Poly-Ubiquitination | Effective for all chain types [24] | Limited for specific chains | Excellent for all chain types after refolding |
| Deubiquitinase Interference | High (requires inhibitors) [34] | Minimal | Minimal |
| Proteasome Interference | High | Minimal | Minimal |
| Reproducibility | Variable due to enzyme activity | High | Extremely high [5] |
Traditional denaturing methods face limitations in UBD-based enrichment because UBDs require native ubiquitin structures for recognition [5]. The DRUSP approach overcomes this by incorporating a refolding step after protein extraction, enabling efficient UBD binding while maintaining the benefits of denaturing conditions for reducing contaminants and enzyme interference [5].
Decision Framework: Selecting the Appropriate Workflow
Diagram 1: Workflow Selection Based on Research Objectives
Key Determinants for Workflow Selection
Research Question Focus: Studies focused on the functional consequences of ubiquitination, including protein-protein interactions and complex formation, should prioritize native workflows. Research aimed specifically at identifying ubiquitination sites and directly modified substrates will benefit from denaturing conditions [24].
Sample Type and Quality: For clinical samples or tissues where maintaining perfect native conditions is challenging, DRUSP provides a robust alternative. For controlled cell culture systems where native interactions can be preserved, traditional native workflows remain valuable [24] [5].
Ubiquitin Chain Type of Interest: Investigations of monoubiquitination benefit from high-affinity UBDs like OtUBD under native conditions [24]. Studies of specific polyubiquitin chain types can be enhanced by combining denaturing conditions with linkage-specific UBDs after refolding [5].
Downstream Applications: Mass spectrometry-based proteomics for ubiquitin site identification requires the specificity of denaturing conditions. Functional biochemical assays may require the preservation of complexes achieved through native purification [24].
Experimental Protocols
Native Workflow for Ubiquitin Interactome Analysis
Principle: This protocol utilizes the high-affinity OtUBD domain under native conditions to capture both covalently ubiquitinated proteins and their non-covalent interaction partners [24].
Step-by-Step Procedure:
Cell Lysis Preparation:
- Prepare native lysis buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100, 1 mM EDTA
- Add fresh protease inhibitors: 1 mM PMSF, 10 μM E-64, 1 μg/mL pepstatin A
- Include deubiquitinase (DUB) inhibitors: 10 mM N-ethylmaleimide (NEM) or 5 mM chloroacetamide (CAA) [34]
- Note: NEM and CAA have different off-target effects; validate for your specific system [34]
Sample Preparation:
- Lyse cells using gentle agitation at 4°C for 30 minutes
- Clarify lysate by centrifugation at 16,000 × g for 15 minutes at 4°C
- Determine protein concentration using Bradford or BCA assay
- Use 1-5 mg total protein per enrichment reaction
OtUBD Affinity Enrichment:
- Pre-equilibrate OtUBD resin with native wash buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1% Triton X-100)
- Incubate clarified lysate with OtUBD resin for 2 hours at 4°C with end-over-end mixing
- Wash resin 3× with native wash buffer
- For stringent washes, include additional washes with high-salt buffer (500 mM NaCl) and no-detergent buffer
Elution and Analysis:
- Elute bound proteins with 2× SDS-PAGE loading buffer containing 100 mM DTT at 95°C for 10 minutes
- Alternatively, for functional studies, elute with competitive elution buffer containing 1-2 mg/mL free ubiquitin
- Analyze by immunoblotting with anti-ubiquitin antibodies or MS-based proteomics
Denaturing Workflow for Covalent Ubiquitinome Profiling
Principle: This protocol utilizes strong denaturants to disrupt non-covalent interactions while preserving covalent ubiquitin modifications, enabling specific isolation of directly ubiquitinated proteins [24].
Step-by-Step Procedure:
Denaturing Lysis Preparation:
- Prepare denaturing lysis buffer: 50 mM Tris-HCl (pH 7.5), 1% SDS, 5 mM EDTA, 10 mM NEM
- Include DUB inhibitors: 10 mM NEM or 5 mM chloroacetamide (CAA)
- Consider adding iodoacetamide (10 mM) for complete cysteine alkylation
Sample Preparation:
- Lyse cells in denaturing buffer with vigorous vortexing
- Heat samples at 95°C for 10 minutes to ensure complete denaturation
- Dilute 10-fold with no-SDS dilution buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100)
- Clarify by centrifugation at 16,000 × g for 15 minutes at 4°C
OtUBD Affinity Enrichment:
- Pre-equilibrate OtUBD resin with denaturing-compatible wash buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 0.1% SDS)
- Incubate diluted lysate with OtUBD resin for 2 hours at 4°C
- Wash resin 3× with denaturing-compatible wash buffer
- Include additional wash with 1 M urea-containing buffer for stringent washing
Elution and Analysis:
- Elute with 2× SDS-PAGE loading buffer at 95°C for 10 minutes
- Process for MS-based proteomics or immunoblot analysis
Advanced Protocol: DRUSP with Tandem Hybrid UBD
Principle: This advanced protocol combines the benefits of denaturing conditions with a refolding step that enables high-efficiency UBD binding, significantly enhancing ubiquitinated protein identification [5].
Step-by-Step Procedure:
Denaturing Extraction:
- Extract proteins using strong denaturing buffer (6 M guanidine hydrochloride or 8 M urea)
- Include DUB inhibitors: 10 mM NEM and 5 mM chloroacetamide
- Heat at 95°C for 10 minutes with shaking
Filter-Based Refolding:
- Dilute denatured extracts with refolding buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100)
- Concentrate using molecular weight cutoff filters
- Perform buffer exchange to remove denaturants gradually
- Final buffer composition: 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5% Triton X-100
Tandem Hybrid UBD Enrichment:
- Use tandem-repeated UBDs or hybrid UBD constructs for enhanced affinity
- Incubate refolded sample with UBD resin for 2 hours at 4°C
- Wash with native wash buffer
- For chain-specific studies, use linkage-specific UBDs [5]
Elution and Processing:
- Elute with competitive elution using free ubiquitin or SDS-PAGE buffer
- Process for downstream applications
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagent Solutions for UBD-Based Ubiquitin Enrichment
| Reagent Category | Specific Examples | Function and Application |
|---|---|---|
| High-Affinity UBDs | OtUBD (from Orientia tsutsugamushi) [24], Tandem Hybrid UBDs [5] | Core affinity reagent for capturing ubiquitinated proteins; high-affinity UBDs enable enrichment of both mono- and polyubiquitinated conjugates |
| Deubiquitinase Inhibitors | N-ethylmaleimide (NEM) [24] [34], Chloroacetamide (CAA) [34] | Preserve ubiquitin signals during processing; choice affects Ub interactor profiles in native pulldowns |
| Denaturing Agents | SDS, Urea, Guanidine hydrochloride [24] [5] | Disrupt non-covalent interactions while preserving covalent ubiquitin modifications |
| Affinity Resins | SulfoLink coupling resin [24], Ni-NTA agarose (for His-tagged UBDs) [24] | Solid support for UBD immobilization |
| Ubiquitin Linkage Tools | Linkage-specific UBDs, Linkage-specific antibodies [19], Chain-specific DUBs (OTUB1 for K48, AMSH for K63) [34] | Enable analysis of specific ubiquitin chain types |
| Detection Antibodies | Anti-ubiquitin (P4D1, E412J) [24], Linkage-specific ubiquitin antibodies [19] | Detect enriched ubiquitinated proteins |
Troubleshooting and Optimization Strategies
Common Challenges and Solutions
Low Ubiquitin Signal Recovery:
- Problem: Insufficient inhibition of deubiquitinases during processing.
- Solution: Implement dual inhibitor approaches (NEM + CAA) and ensure rapid processing. For native workflows, include DUB inhibitors in all buffers [34].
High Background Contamination:
- Problem: Non-specific binding masks specific ubiquitinated proteins.
- Solution: Increase stringency with detergent optimization (Triton X-100 vs. NP-40) and incorporate stepwise salt washes (150-500 mM NaCl) [24].
Incomplete Denaturation or Refolding:
- Problem: Denaturing workflows yield inconsistent results.
- Solution: Validate denaturation efficiency with control proteins and optimize refolding conditions using filter-based methods with gradual denaturant removal [5].
Quality Control Measures
Ubiquitin Chain Integrity Assessment:
- Utilize the UbiCREST method with linkage-specific DUBs (OTUB1 for K48, AMSH for K63) to verify chain composition and integrity [34].
- Include control ubiquitin chains of defined linkage to validate enrichment specificity.
Workflow Performance Validation:
- Implement spike-in controls with defined ubiquitinated standards to quantify enrichment efficiency.
- Compare signal intensity for known ubiquitinated proteins across workflow variations.
The selection between native and denaturing workflows for ubiquitin enrichment represents a critical methodological decision that directly influences experimental outcomes and biological interpretations. Native workflows preserve the physiological context of ubiquitin signaling networks, while denaturing approaches provide specificity for direct ubiquitination events. The emerging DRUSP methodology, which incorporates denaturing extraction followed by refolding, offers a powerful alternative that combines benefits of both approaches, demonstrating substantially improved ubiquitin signal recovery and reproducibility [5].
Researchers should base their workflow selection on clearly defined research objectives, sample characteristics, and downstream applications. The protocols and decision framework presented here provide a foundation for implementing these methods effectively, enabling more robust and comprehensive analysis of the ubiquitinome in health and disease. As ubiquitin biology continues to reveal its complexity, methodological refinements in enrichment strategies will remain essential for deciphering the ubiquitin code and its functional consequences.
Benchmarking UBD Methods: Validation, Comparison, and Best Practices
Within the ubiquitin-proteasome system, the precise enrichment of ubiquitinated proteins is a critical step for understanding the diverse roles of ubiquitin signaling in cellular regulation and disease. The choice of enrichment strategy directly impacts the quality, specificity, and biological relevance of the resulting data. This application note provides a comparative analysis of three fundamental methodologies: Ubiquitin-Binding Domain (UBD)-based affinity enrichment, antibody-based approaches, and Ubiquitin (Ub)-tagging strategies.
Each method offers distinct advantages and limitations in capturing the ubiquitinome, with UBD-based methods emerging as versatile tools for native and denatured ubiquitin chain enrichment. We detail experimental protocols and provide structured data to guide researchers in selecting the optimal approach for specific research contexts within drug development and basic science.
The following table summarizes the core characteristics, advantages, and limitations of the three primary ubiquitin enrichment strategies.
Table 1: Core Characteristics of Ubiquitin Enrichment Strategies
| Feature | UBD-Based Affinity Enrichment | Antibody-Based Enrichment | Ub-Tagging Strategies |
|---|---|---|---|
| Basic Principle | Uses recombinant ubiquitin-binding domains (e.g., OtUBD, TUBEs) to capture ubiquitinated proteins via non-covalent interactions [11] [17]. | Uses anti-ubiquitin antibodies (e.g., P4D1, FK2) or linkage-specific antibodies for immunoprecipitation [11] [47]. | Genetic incorporation of affinity tags (e.g., His, Strep) into ubiquitin for purification [11] [47]. |
| Key Advantage | Captures endogenous ubiquitination; works under native and denaturing conditions; can be linkage-specific [17] [5]. | High specificity for ubiquitin remnants; applicable to tissues without genetic manipulation [11] [47]. | Relatively low-cost and easy to implement in cell culture [11]. |
| Key Limitation | May have lower affinity for monoubiquitination; potential for co-purifying ubiquitin-interacting proteins under native conditions [17] [47]. | High cost; potential sequence bias; cannot distinguish ubiquitination from NEDDylation or ISG15ylation with K-ε-GG antibodies [11] [47]. | Requires genetic manipulation; tagged Ub may not fully mimic endogenous Ub, potentially causing artifacts [11] [47]. |
| Ideal Use Case | System-wide profiling of endogenous ubiquitin chains and interactors; studies requiring specific chain linkage analysis [17] [5]. | Site-specific ubiquitination mapping from clinical samples or animal tissues [11]. | High-throughput substrate screening in engineered cell lines [11]. |
A critical performance differentiator is the ability to handle monoubiquitination versus polyubiquitination. UBDs, particularly Tandem UBDs (TUBEs), exhibit higher affinity for polyubiquitin chains, whereas monoubiquitinated proteins can be more challenging to isolate [17] [47]. In contrast, antibody-based methods that enrich for the K-ε-GG remnant after tryptic digestion are equally effective for mono- and polyubiquitination sites, as they recognize the covalent modification mark rather than the ubiquitin structure itself [47].
Quantitative performance characteristics are summarized in the following table.
Table 2: Quantitative Performance and Practical Considerations
| Aspect | UBD-Based Affinity Enrichment | Antibody-Based Enrichment | Ub-Tagging Strategies |
|---|---|---|---|
| Sensitivity | High for polyubiquitinated proteins; improved by tandem domains (TUBEs) and novel methods like DRUSP [17] [5]. | Very high for modified peptides (e.g., K-ε-GG); can identify >60,000 sites with specialized antibodies [47]. | Moderate; can be impaired by non-specific binding (e.g., histidine-rich proteins) [11]. |
| Specificity | High, but can co-purify non-covalent interactors; specificity for chain types can be engineered [17]. | High for K-ε-GG, though cross-reacts with NEDD8/ISG15; linkage-specific antibodies are available [11] [47]. | Moderate; potential for artifacts from tagged ubiquitin expression [11]. |
| Reproducibility | High, especially with denaturing protocols (e.g., DRUSP) that minimize DUB/protease activity [5]. | High, but can be influenced by lot-to-lot antibody variation. | Generally high within a given engineered system [11]. |
| Throughput | Medium; requires affinity resin preparation and binding steps [17]. | High for proteomic site mapping. | High for screening in cell culture [11]. |
| Cost | Moderate (recombinant protein production) [17]. | High (commercial antibodies) [11]. | Low to Moderate [11]. |
Detailed Experimental Protocols
Protocol 1: UBD-Based Affinity Enrichment using OtUBD
This protocol uses the high-affinity OtUBD from Orientia tsutsugamushi for robust enrichment of ubiquitinated proteins under native or denaturing conditions [17].
Reagents and Materials:
- Plasmids: pRT498-OtUBD or pET21a-cys-His6-OtUBD (Addgene #190089, #190091)
- Affinity Resin: SulfoLink coupling resin
- Lysis Buffers:
- Native Lysis Buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 1 mM EDTA, 10% Glycerol, 1 mM DTT, supplemented with EDTA-free protease inhibitor cocktail, 10 mM N-ethylmaleimide (NEM), and 5 mM iodoacetamide [17].
- Denaturing Lysis Buffer: 50 mM Tris-HCl (pH 7.5), 1% SDS, 1 mM DTT. This is used for the DRUSP method to inactivate DUBs and improve extraction [5].
- Elution Buffer: 1x SDS-PAGE loading buffer containing 100 mM DTT, or 1 M Tris-HCl (pH 6.8) with 1% SDS and 100 mM DTT [17].
Procedure:
- Recombinant OtUBD Purification: Express the His6-tagged OtUBD in E. coli and purify using Ni-NTA affinity chromatography [17].
- OtUBD Resin Preparation: Couple the purified OtUBD protein to SulfoLink resin via cysteine chemistry according to the manufacturer's instructions [17].
- Cell Lysis and Sample Preparation:
- For native conditions: Lyse cells in Native Lysis Buffer. Clarify the lysate by centrifugation at 15,000 × g for 15 min [17].
- For denaturing conditions (DRUSP): Lyse cells in Denaturing Lysis Buffer. Dilute the lysate with a neutral buffer (e.g., 50 mM Tris-HCl, pH 7.5) to reduce SDS concentration, then concentrate and refold proteins using a filter device. The sample is now compatible with the OtUBD resin [5].
- Affinity Enrichment: Incubate the prepared lysate with the OtUBD resin for 2 hours at 4°C with gentle agitation [17].
- Washing: Wash the resin extensively with the appropriate lysis buffer (without inhibitors) to remove non-specifically bound proteins [17].
- Elution: Elute the bound ubiquitinated proteins by incubating the resin with Elution Buffer at 95°C for 10 min [17].
- Downstream Analysis: The eluate can be analyzed by immunoblotting using anti-ubiquitin antibodies or processed for LC-MS/MS analysis [17].
Protocol 2: Antibody-Free Ubiquitination Profiling (AFUP)
AFUP is a chemical proteomics method that selectively enriches endogenous ubiquitination sites without antibodies, avoiding cross-reactivity with NEDD8 or ISG15 [47].
Reagents and Materials:
- Blocking Reagent: Formaldehyde (for dimethylation)
- Deubiquitinases: Recombinant USP2cc (catalytic core) and USP21 [47]
- Labeling Reagent: NHS-SS-Biotin
- Enrichment Resin: Streptavidin Sepharose
- Elution Buffer: 50 mM Tris-HCl (pH 7.5) containing 50 mM DTT or TCEP to cleave the disulfide (SS) bond in NHS-SS-Biotin [47]
Procedure:
- Primary Amine Blocking: Denature the protein extract and block all free amino groups (lysine ε-NH₂ and protein N-terminal α-NH₂) by reductive dimethylation with formaldehyde [47].
- Deubiquitinase Treatment: Digest the sample with a combination of USP2cc and USP21 to specifically remove ubiquitin from modified substrates. This step regenerates the free ε-amine groups only at the former ubiquitination sites [47].
- Clicking Ubiquitination Sites: Label the newly exposed ε-amines by reacting with NHS-SS-Biotin. This reagent selectively tags the ubiquitination sites [47].
- Trypsin Digestion: Digest the biotinylated protein mixture with trypsin to generate peptides [47].
- Streptavidin Enrichment: Incubate the tryptic peptides with Streptavidin Sepharose to capture the biotinylated peptides (which correspond to ubiquitination sites) [47].
- Washing and Elution: After extensive washing, elute the captured peptides by reducing the disulfide bond in the biotin linker with DTT or TCEP [47].
- LC-MS/MS Analysis: Identify the site-specific ubiquitination by analyzing the eluted peptides via Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) [47].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Ubiquitin Enrichment
| Reagent / Tool | Function / Description | Example Use Cases |
|---|---|---|
| OtUBD [17] | High-affinity ubiquitin-binding domain from O. tsutsugamushi (Kd in low nM range). | Core component for UBD-based affinity resins; enrichment of mono- and polyubiquitinated proteins. |
| Tandem Hybrid UBD (ThUBD) [5] | Engineered UBD with multiple domains to enhance avidity for polyubiquitin chains. | Combined with DRUSP method for high-efficiency, unbiased ubiquitinome profiling. |
| K-ε-GG Antibody [47] | Antibody recognizing diglycine remnant on lysine after trypsin digestion of ubiquitinated proteins. | Gold standard for proteome-wide mapping of ubiquitination sites by MS. |
| UbiSite Antibody [47] | Antibody recognizing C-terminal 13 aa of Ub after Lys-C digestion. | Alternative site-specific enrichment; avoids cross-reactivity with NEDD8/ISG15. |
| Linkage-Specific Ub Antibodies [11] | Antibodies specific to M1, K48, K63, etc., linkages. | Immunoblotting or enrichment of proteins modified with specific ubiquitin chain types. |
| N-Ethylmaleimide (NEM) [17] | Cysteine protease inhibitor. | Essential in lysis buffers to inhibit deubiquitinating enzymes (DUBs) and preserve ubiquitin signals. |
| USP2cc / USP21 [47] | Catalytic core domains of deubiquitinating enzymes with broad linkage specificity. | Generation of free amines in the AFUP protocol; ubiquitin chain restriction analysis. |
The strategic selection of an ubiquitin enrichment method is paramount for successful research outcomes. UBD-based affinity methods, particularly when combined with novel sample preparation techniques like DRUSP, offer powerful, versatile, and economical tools for system-wide analysis of endogenous ubiquitination. Antibody-based approaches remain the gold standard for high-sensitivity, site-specific mapping, especially in clinical samples. Ub-tagging provides a straightforward method for substrate identification in genetically tractable systems.
Researchers should base their selection on the specific biological question, sample type, and required output—whether it is global ubiquitinome profiling, site-specific quantification, or the analysis of specific ubiquitin chain linkages. A combination of these methods often provides the most comprehensive insights into the complex landscape of ubiquitin signaling.
Ubiquitin-binding domain (UBD)-based affinity enrichment represents a cornerstone technique in proteomics for the isolation and identification of ubiquitinated proteins, a process critical for understanding diverse cellular signaling pathways [19]. The versatility of ubiquitination, encompassing various chain topologies and linkages, necessitates enrichment tools that are not only highly affine but also specific and reproducible [51]. While multiple UBDs have been developed, their performance must be rigorously quantified using standardized metrics to ensure data quality and biological relevance. This application note provides a detailed protocol and a comprehensive set of performance metrics for evaluating UBD-based affinity enrichment methods, focusing on the high-affinity OtUBD and ThUBD systems. We present quantitative data on their efficiency, specificity, and reproducibility, alongside a standardized experimental workflow to guide researchers in the robust characterization of ubiquitinated proteomes.
Key Performance Metrics for UBD-Based Enrichment
The evaluation of any UBD-based enrichment method rests on three fundamental pillars: its efficiency in capturing the target ubiquitin signal, its specificity in distinguishing this signal from non-specific background, and the reproducibility of the process across technical and biological replicates. The following table summarizes core quantitative metrics used for this assessment, with benchmark values from recent literature for the OtUBD and ThUBD systems.
Table 1: Key Performance Metrics for UBD-Based Enrichment Methods
| Metric | Description | Benchmark Data (Method) |
|---|---|---|
| Enrichment Efficiency | Strength of ubiquitin signal recovered post-enrichment compared to input. | ~10-fold increase in ubiquitin signal (DRUSP-ThUBD) [5]; Specific binding of ~5 pmol of polyUb chains (ThUBD 96-well plate) [1]. |
| Signal-to-Noise Ratio | Ratio of ubiquitinated protein signal to co-purifying non-ubiquitinated proteins. | Demonstrated by strong ubiquitin smears in immunoblots with minimal background; use of denaturing conditions in OtUBD protocol to minimize non-covalent interactors [24]. |
| Ubiquitin Chain Linkage Bias | Ability to enrich various ubiquitin chain linkages (K48, K63, etc.) without preference. | ThUBD shows "no bias toward any type of ubiquitin chain" [1]; OtUBD resin enriches both mono- and poly-ubiquitinated proteins [24]. |
| Reproducibility (Quantitative) | Coefficient of variation (CV) for ubiquitinated protein identification across replicates. | DRUSP-ThUBD demonstrates "enhanced quantitative accuracy and reproducibility for ubiquitinomics" [5]. |
| Dynamic Range | Ability to ubiquitinated proteins across a wide abundance range. | Successful enrichment from complex mammalian cell lysates; identification of hundreds to thousands of ubiquitination sites via LC-MS/MS [24] [19]. |
Featured Reagent Solutions
The success of UBD-based enrichment is fundamentally linked to the quality and properties of the reagents employed. The table below catalogs essential tools, highlighting the unique advantages of specific UBDs that have been systematically characterized.
Table 2: Essential Research Reagents for UBD-Based Affinity Enrichment
| Reagent / Tool | Function / Description | Key Feature / Application |
|---|---|---|
| OtUBD Affinity Resin [24] | High-affinity UBD from O. tsutsugamushi coupled to a solid support. | High-affinity (Kd in low nM for monoubiquitin [25]); Enriches both mono- and poly-ubiquitinated proteins; Offers native and denaturing workflows. |
| ThUBD-Coated Plates [1] | Tandem hybrid UBD coated onto high-density 96-well plates. | High-throughput, unbiased recognition of ubiquitin chains; Enables high-sensitivity, plate-based quantification of ubiquitination. |
| DRUSP Protocol [5] | Denatured-Refolded Ubiquitinated Sample Preparation method. | Enhances ubiquitin signal and extraction; Couples with UBDs (e.g., ThUBD) for improved robustness and reproducibility in ubiquitinomics. |
| Linkage-Specific DiUb Probes [51] | Chemically synthesized diubiquitin (e.g., via click chemistry) of defined linkage. | Serves as a standard for evaluating linkage-specific enrichment bias of UBDs; Resistant to deubiquitinase (DUB) activity. |
| Anti-Ubiquitin Antibodies [19] | Antibodies (e.g., P4D1, FK2) for immunoblotting validation. | Critical for downstream validation of enrichment efficiency and specificity via Western blot. |
Experimental Protocol: OtUBD-Based Enrichment and Performance Assessment
This section provides a detailed step-by-step protocol for enriching ubiquitinated proteins from mammalian cell lysates using the OtUBD affinity resin, incorporating steps to evaluate key performance metrics.
Materials and Reagents
- OtUBD Affinity Resin: Prepared as described [24] using the pET21a-cys-His6-OtUBD plasmid (Addgene #190091) coupled to SulfoLink resin.
- Cell Lysis Buffers:
- Native Lysis Buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 1 mM DTT, 10 mM N-ethylmaleimide (NEM), cOmplete EDTA-free protease inhibitor cocktail.
- Denaturing Lysis Buffer: 50 mM Tris-HCl (pH 7.5), 1% SDS, 10 mM NEM, protease inhibitors.
- Wash Buffers: Native Wash Buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1% Triton X-100); High-Salt Wash Buffer (50 mM Tris-HCl pH 7.5, 500 mM NaCl, 0.1% Triton X-100).
- Elution Buffer: 1X SDS-PAGE loading buffer containing 50 mM DTT.
- Antibodies: Mouse anti-ubiquitin P4D1 (Enzo) and HRP-linked anti-mouse secondary antibody.
Step-by-Step Procedure
Cell Lysis and Protein Extraction:
- Harvest HEK293T cells and lyse in either Native or Denaturing Lysis Buffer.
- For denaturing lysis, sonicate the lysate and boil for 5 minutes. Dilute the SDS concentration to 0.1% before proceeding.
- Clear the lysate by centrifugation at 16,000 × g for 15 min at 4°C. Quantify the protein concentration.
Affinity Enrichment Pulldown:
- Incubate 1-2 mg of total cell lysate with 50 μL of settled OtUBD affinity resin for 2 hours at 4°C with end-over-end mixing.
- Pellet the resin by brief centrifugation and carefully remove the supernatant (the "Flow-Through" fraction).
Stringent Washing:
- Wash the resin three times with 1 mL of Native Wash Buffer.
- Perform two additional washes with 1 mL of High-Salt Wash Buffer.
- Note: Retain aliquots of the input lysate, flow-through, and first wash for downstream immunoblot analysis.
Elution of Ubiquitinated Proteins:
- Elute the bound proteins by adding 50 μL of Elution Buffer to the resin and heating at 95°C for 10 minutes.
- Centrifuge and collect the eluate.
Downstream Analysis:
- Analyze the Input, Flow-Through, Wash, and Elution fractions by SDS-PAGE and immunoblotting with anti-ubiquitin antibody.
- For proteomic analysis, process the eluted proteins for LC-MS/MS.
Performance Assessment Workflow
The following diagram visualizes the key steps for conducting the enrichment and calculating the critical performance metrics.
Diagram 1: Performance assessment workflow for UBD-based enrichment.
Data Analysis and Interpretation
Calculating Key Metrics
- Enrichment Efficiency: Compare the intensity of the ubiquitin signal in the elution fraction to that in the input lysate via immunoblot densitometry or by the number of unique ubiquitination sites identified by LC-MS/MS. A successful enrichment should show a strong, high-molecular-weight smear in the eluate that is markedly enriched over the input.
- Specificity: Inspect the wash and elution fractions by immunoblotting. Specific enrichment is indicated by a clear ubiquitin signal in the eluate with minimal to no signal in the final wash fraction, indicating effective removal of non-specifically bound proteins.
- Reproducibility: Process at least three independent biological replicates through the entire workflow. For LC-MS/MS data, calculate the coefficient of variation (CV) for the number of identified ubiquitinated proteins or ubiquitination sites across replicates. A CV of less than 20% is typically indicative of high reproducibility.
Troubleshooting Common Issues
- High Background in Eluate: Increase the number of washes or incorporate a high-salt wash step to disrupt non-specific ionic interactions.
- Weak Ubiquitin Signal: Verify resin activity and binding capacity. Ensure fresh NEM is added to lysis buffers to inhibit deubiquitinases. For deep ubiquitinome coverage, consider using the DRUSP protocol to improve protein extraction and refolding prior to enrichment [5].
- Inconsistent Results Between Replicates: Standardize all incubation times, wash volumes, and resin handling procedures. Ensure consistent protein input amounts across replicates.
Robust assessment of enrichment efficiency, specificity, and reproducibility is paramount for generating high-quality data in ubiquitin proteomics. The high-affinity OtUBD and ThUBD systems, characterized by their strong binding and low linkage bias, provide a solid foundation for such studies. By adhering to the detailed protocols and quantitative metrics outlined in this application note, researchers can critically evaluate and optimize their UBD-based enrichment strategies, thereby ensuring the reliability and biological significance of their findings in the complex landscape of ubiquitin signaling.
Protein ubiquitination is a fundamental post-translational modification that regulates nearly all eukaryotic cellular processes, including proteasome-mediated degradation, DNA repair, cell signaling, and endocytosis [27] [19]. The Ubiquitin Binding Domain (UBD)-based affinity enrichment methods have emerged as powerful tools for investigating the "ubiquitin code." However, the complexity of ubiquitin signaling—encompassing monoubiquitination, multiple monoubiquitination, and polyubiquitin chains of eight distinct homotypic linkages and heterotypic/branched architectures—presents significant challenges for accurate characterization [27] [19]. Furthermore, the typically low stoichiometry of modified species and the dynamic nature of ubiquitination necessitate implementing rigorous cross-validation strategies. This Application Note details a multidisciplinary framework for confirming ubiquitination findings, ensuring research robustness and reproducibility within UBD-based affinity enrichment studies.
Orthogonal Biochemical Methods for Validation
UBD-Based Affinity Enrichment with Controlled Stringency
The OtUBD (from Orientia tsutsugamushi deubiquitinase) affinity resin represents a significant advancement in UBD-based tools due to its high affinity for ubiquitin in the low nanomolar range [29]. Its key application for cross-validation lies in employing different buffer formulations to distinguish covalently ubiquitinated proteins from associated interactors.
- Denaturing Workflow: Using strong denaturants (e.g., 8 M urea) disrupts non-covalent protein-protein interactions. This allows for the specific isolation of directly ubiquitinated proteins, providing high confidence in identified substrates [4] [29].
- Native Workflow: Performed under physiological or mild conditions, this workflow co-purifies both covalently ubiquitinated proteins and proteins that interact with ubiquitin or ubiquitinated substrates (the ubiquitin interactome) [17] [24].
Comparing results from denaturing versus native OtUBD enrichments from the same biological sample allows researchers to validate true ubiquitin conjugates and simultaneously gain insight into potential functional complexes [29].
Immunoblotting with Linkage-Specific Antibodies
Following enrichment, immunoblotting remains a cornerstone for validation.
- Molecular Weight Shift: The attachment of ubiquitin, especially polyubiquitin chains, causes a characteristic increase in apparent molecular weight, visible as a ladder or smear on a Western blot [52].
- Antibody Confirmation: Using antibodies against ubiquitin (e.g., P4D1) or epitope tags on expressed ubiquitin confirms the presence of ubiquitin in enriched samples [52] [17]. The development of linkage-specific antibodies (e.g., for K48 or K63 chains) further allows for the determination of ubiquitin chain topology, providing functional context to the modification [19].
Table 1: Orthogonal Biochemical Methods for Ubiquitination Validation
| Method | Key Principle | Key Advantage | Common Application in Cross-Validation |
|---|---|---|---|
| OtUBD (Denaturing) | Enrichment under denaturing conditions (e.g., 8M Urea) | Isolates covalently ubiquitinated proteins, minimizing co-purifying interactors [4] [17] | Primary validation of ubiquitinated substrates from complex lysates |
| OtUBD (Native) | Enrichment under native/non-denaturing conditions | Co-purifies ubiquitinated proteins and their interacting partners [24] [29] | Mapping the ubiquitin interactome; contrast with denaturing results |
| Western Blot (Molecular Weight) | Detection of increased molecular weight on SDS-PAGE | Simple, readily accessible; indicates mono vs. polyubiquitination [52] | Post-enrichment confirmation of successful ubiquitin pull-down |
| Linkage-Specific Antibodies | Immunoblot with antibodies specific to ubiquitin linkage types | Defines chain topology (e.g., K48 vs K63), providing functional insight [19] | Characterizing the nature of the ubiquitin signal detected |
Diagram 1: Workflow for cross-validation using OtUBD enrichment under different stringency conditions combined with orthogonal confirmation methods.
Computational and Mass Spectrometry Approaches
Virtual Western Blot Analysis
A powerful computational method for high-throughput validation involves reconstructing "virtual Western blots" from liquid chromatography-tandem mass spectrometry (LC-MS/MS) data generated from gel-separated samples (1D geLC-MS/MS) [52].
- Principle: The experimental molecular weight of a protein identified by MS is computed based on its distribution across gel fractions using Gaussian curve fitting. This value is compared to its theoretical molecular weight [52].
- Validation Criteria: True ubiquitin conjugates exhibit a significant increase in experimental molecular weight compared to their theoretical mass, consistent with the addition of ubiquitin (∼8.5 kDa for monoubiquitination) or polyubiquitin chains. Applying stringent thresholds based on ubiquitin mass and experimental variation can achieve an estimated false discovery rate of ∼8% [52].
- Utility: This method is particularly effective for validating large-scale datasets where traditional Western blotting for hundreds of candidates is impractical. It has been shown that about 95% of proteins with identified ubiquitination sites display a convincing molecular weight shift on virtual Western blots [52].
Direct Identification of Ubiquitination Sites via diGly Antibody Enrichment
A gold standard for confirming ubiquitination is the direct MS-based mapping of modification sites, typically following tryptic digestion.
- diGly Remnant: Trypsin cleavage of ubiquitinated proteins leaves a characteristic di-glycine (GlyGly) remnant on modified lysine residues, resulting in a mass shift of 114.0429 Da [52] [19].
- Antibody Enrichment: Antibodies specifically recognizing this diGly-ε-Lys motif are used to enrich modified peptides from complex digests, dramatically improving the sensitivity of site identification [19] [29].
- Cross-Validation: The identification of a diGly-modified lysine residue on a protein candidate from an OtUBD enrichment provides definitive evidence of its ubiquitination. This method directly complements the protein-level enrichment achieved by UBD-based approaches [29].
Table 2: Mass Spectrometry and Computational Validation Techniques
| Method | Key Principle | Key Advantage | Considerations |
|---|---|---|---|
| Virtual Western Blot | Computational MW determination from geLC-MS/MS data | High-throughput validation for large candidate lists [52] | Effective for proteins >100 kDa; requires gel separation pre-MS |
| diGly Remnant MS (Bottom-Up) | Enrichment of peptides with GlyGly modification on Lys | Direct, site-specific confirmation of ubiquitination [52] [19] | Cannot detect non-lysine ubiquitination (Ser/Thr/Cys) |
| Ubiquitin Site Predictors (e.g., UbPred) | Machine learning prediction of ubiquitination sites based on sequence motifs | Guides experimental design and prioritization of lysine mutants for validation [53] | Predictive only; requires experimental confirmation |
Detailed Experimental Protocol
Cross-Validation Workflow Using OtUBD Enrichment
This protocol outlines the steps for validating ubiquitinated proteins from yeast or mammalian cell lysates using OtUBD, followed by cross-validation with virtual Western blot and diGly remnant analysis.
Part A: OtUBD Affinity Enrichment under Denaturing Conditions
Cell Lysis and Denaturation:
- Resuspend cell pellets in denaturing lysis buffer (e.g., 10 mM Tris-HCl, pH 8.0, 0.1 M NaH₂PO₄, 8 M urea, 10 mM β-mercaptoethanol) [52].
- Clarify the lysate by centrifugation at high speed (e.g., 70,000 × g for 30 min) to remove insoluble debris.
Affinity Purification:
- Incubate the clarified lysate with OtUBD affinity resin for 1-2 hours at room temperature with gentle agitation [17] [24].
- Wash the resin extensively with denaturing wash buffer (e.g., lysis buffer with adjusted pH and potentially 0.2% SDS) to remove non-specifically bound proteins [52] [29].
- Elute the bound ubiquitinated proteins using a low-pH elution buffer (e.g., 50 mM glycine, pH 2.5) or by boiling in SDS-PAGE sample buffer [24].
Part B: Cross-Validation of Enriched Proteins
Virtual Western Blot Analysis:
- Resolve a portion of the eluate on a 6-12% gradient SDS-PAGE gel. Cut the entire gel lane into multiple slices (e.g., 10-40 bands) [52].
- Perform in-gel trypsin digestion on each gel band [52].
- Analyze the resulting peptides by LC-MS/MS. Search the data against the appropriate protein database, allowing for the dynamic modification of lysine with the GlyGly remnant (+114.0429 Da).
- Compute the experimental molecular weight for each identified protein based on the distribution of its spectral counts across the gel fractions. Accept proteins as validated ubiquitin conjugates only if their experimental MW significantly exceeds their theoretical MW (incorporating thresholds for the mass of ubiquitin) [52].
diGly Remnant Enrichment and Site Mapping:
- Take another portion of the OtUBD eluate and digest the proteins in solution with trypsin.
- Use anti-diGly (GlyGly-ε-Lys) antibodies to immunoprecipitate the modified peptides from the digested mixture [19].
- Analyze the enriched peptides by LC-MS/MS to identify the specific sites of ubiquitination on the candidate proteins.
Diagram 2: Detailed experimental workflow for the cross-validation of ubiquitinated proteins, combining OtUBD enrichment with virtual Western blot analysis and diGly remnant site mapping.
Research Reagent Solutions
Table 3: Essential Research Reagents for UBD-Based Ubiquitin Enrichment and Validation
| Reagent / Tool | Function / Application | Key Feature / Rationale |
|---|---|---|
| OtUBD Affinity Resin [4] [29] | High-affinity enrichment of mono- and polyubiquitinated proteins from lysates. | High nanomolar affinity, works on all chain types and non-canonical sites; compatible with denaturing conditions. |
| Denaturing Lysis Buffer (8M Urea) [52] | Cell lysis and protein denaturation prior to enrichment. | Disrupts non-covalent interactions, ensuring isolation of covalently ubiquitinated proteins. |
| Anti-diGly Remnant Antibodies [19] | Immunoaffinity enrichment of ubiquitinated peptides for LC-MS/MS site mapping. | Provides definitive, site-specific evidence of ubiquitination on lysine residues. |
| Linkage-Specific Ubiquitin Antibodies [19] | Immunoblotting to determine ubiquitin chain topology. | Identifies specific chain linkages (e.g., K48, K63), offering functional insights. |
| Tandem Ubiquitin-Binding Entities (TUBEs) [19] | Alternative enrichment tool, particularly for polyubiquitinated proteins. | Protects polyubiquitin chains from deubiquitinases (DUBs) during purification. |
| Epitope-Tagged Ubiquitin (e.g., His-, Strep-) [19] | Ectopic expression for affinity purification of ubiquitinated proteome. | Allows purification under fully denaturing conditions; may alter endogenous ubiquitination dynamics. |
The complexity of the ubiquitin code demands a rigorous, multi-faceted approach to experimental validation. Relying on a single method introduces the risk of false positives from non-specifically bound proteins or the misinterpretation of ubiquitin-interacting proteins as true conjugates. The integrated strategy detailed herein—combining the high-affinity, flexible enrichment capabilities of OtUBD under controlled stringency with the computational power of virtual Western blots and the definitive site-specific evidence from diGly remnant mass spectrometry—creates a robust framework for confirmation. By adopting these cross-validation techniques, researchers in drug discovery and basic science can generate highly reliable ubiquitin proteome datasets, accelerating our understanding of ubiquitin biology and the development of targeted therapeutics.
Ubiquitination is a critical post-translational modification that regulates diverse cellular functions, from protein degradation to DNA repair and immune signaling [54] [19]. The versatility of ubiquitin signaling stems from the complexity of ubiquitin conjugates, which can range from a single ubiquitin monomer to polymers of different lengths and linkage types [19]. This diversity, often referred to as the "ubiquitin code," presents significant challenges for comprehensive analysis. Ubiquitin-binding domains (UBDs) have emerged as powerful tools for enriching ubiquitinated proteins, but traditional UBD-based methods suffer from critical limitations including linkage bias, affinity restrictions, and inadequate coverage of heterogeneous chains [1] [4] [19]. This application note examines these limitations within the context of ubiquitin research and presents advanced methodologies designed to overcome these challenges, enabling more precise and comprehensive characterization of the ubiquitinome.
Current Limitations in UBD-Based Affinity Enrichment
Technical Challenges and Methodological Gaps
Traditional UBD-based approaches face several inherent limitations that compromise their effectiveness in ubiquitin research. The table below summarizes the primary technical challenges and their implications for research outcomes.
Table 1: Key Limitations of Conventional UBD-Based Enrichment Methods
| Limitation | Impact on Research | Common Affected Applications |
|---|---|---|
| Linkage Bias | Incomplete ubiquitinome profiling; skewed biological interpretations [1] | Proteomic studies, signaling pathway analysis |
| Low Affinity for Mono-Ubiquitination | Poor recovery of monoubiquitinated substrates [24] | Endocytosis, DNA repair, histone regulation studies |
| Inadequate Chain Length Coverage | Limited detection of specific polyubiquitin chain architectures [1] | Proteasomal degradation, autophagy research |
| Inability to Resolve Heterogeneous Chains | Failure to characterize mixed/branched ubiquitin chains [19] | Complex signaling nodes, therapeutic target validation |
Specific Deficiencies in Commonly Used UBDs
The tandem ubiquitin-binding entities (TUBEs) widely used in many commercial assays exhibit inherent biases that limit their application. These tools demonstrate low affinity for ubiquitin chains and show preferential binding to specific ubiquitin chain types, potentially yielding results that do not accurately reflect the true intracellular ubiquitination status [1]. Furthermore, TUBEs perform poorly against monoubiquitinated proteins, which often constitute a substantial fraction of ubiquitinated proteins in mammalian cells and tissues [24]. This coverage gap is particularly problematic for researchers investigating processes primarily regulated by monoubiquitination, such as endocytic trafficking and epigenetic regulation.
Advanced UBD Technologies for Comprehensive Ubiquitinome Profiling
Engineered Tandem Hybrid UBDs (ThUBDs)
The ThUBD platform represents a significant advancement in UBD technology, engineered to overcome the linkage bias of earlier methods. By combining the advantages of different ubiquitin-binding domains, ThUBD exhibits unbiased recognition of various ubiquitin chain types while maintaining high affinity for polyubiquitinated proteins [1]. This technology has been adapted into high-density 96-well plates capable of capturing approximately 5 pmol of polyubiquitin chains when coated with 1.03 μg ± 0.002 of ThUBD, enabling high-throughput screening applications for drug discovery programs targeting the ubiquitin-proteasome system [1].
Table 2: Performance Comparison of UBD Technologies
| Parameter | Traditional TUBEs | ThUBD Platform | OtUBD Technology |
|---|---|---|---|
| Affinity Range | Low nanomolar (biased) [1] | High affinity (unbiased) [1] | ~5 nM Kd (monoubiquitin) [25] |
| Monoubiquitin Detection | Limited [24] | Efficient [1] | Excellent [4] [24] |
| Polyubiquitin Chain Preference | Linkage-dependent [1] | Linkage-independent [1] | Chain length-dependent (>3 ubiquitins) [25] |
| Proteomic Coverage | Partial ubiquitinome | Comprehensive ubiquitinome | Broad (mono- and polyubiquitin) [4] |
| Throughput Capability | Moderate | High (96-well format) [1] | Low to moderate |
High-Affinity OtUBD from Orientia tsutsugamushi
The ubiquitin-binding domain derived from Orientia tsutsugamushi (OtUBD) represents a breakthrough in affinity performance, binding monoubiquitin with an unprecedented dissociation constant of approximately 5 nM [25]. This remarkable affinity stems from a unique structural transition wherein the UBD changes from a poorly folded to well-ordered state upon ubiquitin binding [25]. The OtUBD affinity resin can strongly enrich both mono- and poly-ubiquitinated proteins from crude lysates, addressing a critical coverage gap in conventional methods [4] [24].
Diagram 1: OtUBD-Ubiquitin Binding Mechanism
Experimental Protocols for Enhanced Ubiquitin Enrichment
ThUBD-Coated Plate Protocol for High-Throughput Screening
Principle: This method leverages the unbiased binding characteristics of ThUBD in a standardized 96-well format for specific, rapid, and precise detection of protein ubiquitination [1].
Materials:
- ThUBD-coated Corning 3603-type 96-well plates
- Binding buffer: 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.1% Triton X-100
- Wash buffer: 50 mM Tris-HCl (pH 8.0), 500 mM NaCl, 0.5% Triton X-100
- Detection reagent: ThUBD-HRP conjugate
- Blocking solution: 5% non-fat milk in TBST
Procedure:
- Coating: Confirm ThUBD coating density of 1.03 μg ± 0.002 per well
- Blocking: Block plates with 5% non-fat milk for 1 hour at room temperature
- Sample Incubation: Add cell lysates containing 50-100 μg total protein to each well
- Binding: Incubate for 2 hours at 4°C with gentle agitation
- Washing: Perform three washes with high-stringency wash buffer
- Detection: Incubate with ThUBD-HRP conjugate for 1 hour
- Signal Development: Add chemiluminescent substrate and measure signal
Validation: The method specifically detects polyubiquitin chains with a detection limit of approximately 5 pmol, without cross-reactivity to SUMO or NEDD8 [1].
OtUBD Affinity Resin Protocol for Ubiquitinome Analysis
Principle: This protocol utilizes the exceptional affinity of OtUBD for comprehensive enrichment of ubiquitinated proteins from complex biological samples [4] [24].
Materials:
- OtUBD affinity resin (prepared as described in [24])
- Lysis buffer (native): 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 10% glycerol
- Lysis buffer (denaturing): 50 mM Tris-HCl (pH 7.5), 1% SDS, 1 mM N-ethylmaleimide (NEM)
- Elution buffer: 50 mM Tris-HCl (pH 7.5), 2% SDS, 10 mM DTT
- Protease inhibitor cocktail (EDTA-free)
- DUB inhibitors (NEM or PR-619)
Procedure:
- Cell Lysis:
- For native conditions: Use NP-40-based lysis buffer to preserve protein interactions
- For denaturing conditions: Use SDS-based buffer to isolate covalently ubiquitinated proteins
Resin Preparation:
- Equilibrate OtUBD resin with appropriate binding buffer
- Use 20-50 μL resin per mg of total protein
Enrichment:
- Incubate clarified lysate with resin for 2-4 hours at 4°C
- Use end-over-end rotation for maximum binding efficiency
Washing:
- Perform sequential washes with binding buffer containing 0.1% Triton X-100
- Include a high-salt wash (500 mM NaCl) to reduce non-specific binding
Elution:
- Elute with SDS-PAGE loading buffer containing DTT for downstream immunoblotting
- For proteomics, elute with 2% SDS or competitive elution with free ubiquitin
Applications: This protocol has been successfully used for immunoblotting, differential proteomics, and UbiCREST (ubiquitin chain restriction) assays, working with all types of ubiquitin conjugates [4] [24].
Research Reagent Solutions for Ubiquitin Studies
Table 3: Essential Reagents for Advanced Ubiquitin Enrichment Studies
| Reagent/Category | Specific Examples | Function/Application | Considerations |
|---|---|---|---|
| High-Affinity UBDs | ThUBD, OtUBD [1] [4] | Unbiased ubiquitin enrichment | OtUBD requires expression and purification |
| Affinity Resins | SulfoLink coupling resin [24] | UBD immobilization | Covalent coupling preserves binding capacity |
| Cell Lysis Reagents | N-ethylmaleimide (NEM) [24] | DUB inhibition | Critical for preserving ubiquitin signals |
| Protease Inhibitors | EDTA-free cocktails [24] | Prevent protein degradation | EDTA interferes with certain UBDs |
| Elution Reagents | SDS/DTT, free ubiquitin [24] | Sample recovery | Competitive elution preserves protein interactions |
| Detection Antibodies | Linkage-specific anti-ubiquitin [19] | Chain-type characterization | Limited to characterized linkage types |
Analytical Framework for Method Selection
Diagram 2: UBD Method Selection Framework
The limitations of traditional UBD-based methods in addressing artifacts, coverage gaps, and heterogeneous chains represent significant challenges in ubiquitin research. Advanced technologies like ThUBD and OtUBD provide powerful solutions to these problems, offering unbiased recognition, exceptional affinity, and comprehensive coverage of diverse ubiquitin modifications. The experimental frameworks presented herein enable researchers to select appropriate methodologies based on their specific research questions, whether focused on monoubiquitination, specific chain types, or exploratory ubiquitinome profiling. As the ubiquitin field continues to evolve, these advanced UBD technologies will play an increasingly crucial role in deciphering the complex ubiquitin code and developing targeted therapeutic interventions for diseases characterized by ubiquitination dysregulation.
Ubiquitin Binding Domain (UBD)-based affinity enrichment represents a cornerstone technique in modern proteomics for the systematic analysis of protein ubiquitination. The ubiquitin-proteasome system (UPS) serves as the core machinery for targeted protein degradation and quality control in eukaryotes, playing a pivotal role in maintaining proteostasis and cellular homeostasis [1]. Beyond protein degradation, the UPS orchestrates nearly all cellular processes, including DNA repair, cell cycle regulation, and immune responses [1]. Its dysregulation is intimately linked to the pathogenesis of prevalent human diseases, including cancers and neurodegenerative disorders [1].
The diversity in ubiquitin chain topologies and linkages allows a corresponding diversity in substrate protein fates across numerous cellular pathways [24]. Traditional methodologies for detecting ubiquitination signals, including mass spectrometry-based analysis, antibody-based immunological assays, and tandem ubiquitin-binding entities (TUBEs), each present significant limitations [24] [1]. These include spurious ubiquitination patterns from epitope-tagged ubiquitin overexpression, insufficient sensitivity or specificity of anti-ubiquitin antibodies, and poor efficiency against monoubiquitinated proteins with TUBEs [24]. The development of high-affinity UBDs like OtUBD and ThUBD has revolutionized this field by enabling highly efficient, unbiased enrichment of both mono- and polyubiquitinated proteins from complex biological samples [24] [1].
Comparative Analysis of UBD Technologies
OtUBD: High-Affinity Bacterial Derived Domain
OtUBD is a high-affinity ubiquitin-binding domain derived from a large deubiquitinase protein (OtDUB) from the bacterial pathogen Orientia tsutsugamushi [24]. This domain exhibits exceptional affinity for ubiquitin, with a dissociation constant in the low nanomolar range, prompting its development into an affinity resin for enriching ubiquitinated proteins from complex biological samples [24]. The OtUBD system provides researchers with a versatile and economical tool for comprehensive study of ubiquitin biology, supporting downstream applications including immunoblotting, differential proteomics, and UbiCREST (ubiquitin chain restriction) applications [24].
Key advantages of OtUBD include:
- Capability to strongly enrich both mono- and polyubiquitinated proteins from crude lysates
- Compatibility with both native and denaturing workflow conditions
- Proven effectiveness with budding yeast and mammalian cell lysates, with adaptability to other biological samples and organisms
- Ability to distinguish covalently ubiquitinated proteins (the ubiquitinome) from ubiquitin- or ubiquitinated protein-interacting proteins (the ubiquitin interactome)
ThUBD: Engineered Tandem Hybrid Domain
ThUBD represents an engineered tandem hybrid ubiquitin-binding domain designed to overcome limitations of previous TUBE technologies [1]. This laboratory-developed domain combines the advantages of different ubiquitin-binding domains, showing not only high affinity for polyubiquitinated proteins but also exhibiting no bias toward any type of ubiquitin chain, enabling specific and sensitive detection of ubiquitinated proteins [1]. The ThUBD technology has been further developed into highly sensitive and rapid detection platforms including TUF-WB, TUF-WB+, and high-density 96-well plate assays for high-throughput screening applications [1].
Comparative performance characteristics:
- ThUBD coated on 96-well plates enables specific binding to approximately 5 pmol of polyubiquitin chains
- Demonstrates unbiased enrichment and detection of different ubiquitin chains
- Provides enhanced sensitivity compared to TUBE-based platforms
- Enables high-throughput detection of protein ubiquitination signals and batch detection of target protein ubiquitination in samples
Technical Comparison of UBD Platforms
Table 1: Comparative Analysis of UBD Affinity Enrichment Platforms
| Feature | OtUBD | ThUBD | Traditional TUBEs |
|---|---|---|---|
| Affinity Constant | Low nanomolar range [24] | High affinity (specific values not published) [1] | Low affinity [1] |
| Monoubiquitin Enrichment | Strong enrichment [24] | Not specified | Poor efficiency [24] |
| Polyubiquitin Enrichment | Strong enrichment [24] | High affinity enrichment [1] | Highly efficient [24] |
| Linkage Bias | Works with all types of ubiquitin conjugates [24] | Unbiased recognition of different ubiquitin chains [1] | Bias toward specific ubiquitin chain types [1] |
| Throughput Capability | Standard proteomics workflow | High-throughput 96-well plate format [1] | Variable |
| Cost Effectiveness | Economical tool [24] | Not specified | Variable commercial costs |
Experimental Protocols for UBD-Based Ubiquitinome Analysis
OtUBD Affinity Resin Preparation
Principle: Recombinant OtUBD protein is expressed and purified for covalent coupling to solid support matrix, creating a high-affinity enrichment resin for ubiquitinated proteins.
Materials:
- Plasmids: pRT498-OtUBD (Addgene, plasmid #190089) or pET21a-cys-His6-OtUBD (Addgene, plasmid #190091) [24]
- Expression host: BL21(DE3) or similar E. coli strains
- Chromatography resin: SulfoLink coupling resin (Thermo Scientific, catalog number: 20402) [24]
- Purification reagents: Ni-NTA agarose (Qiagen, catalog number: 30230), imidazole, IPTG [24]
- Buffers: Luria-Bertani (LB) medium, phosphate-buffered saline (DPBS), elution buffers [24]
Procedure:
- Transform appropriate expression plasmid into BL21(DE3) E. coli cells and plate on LB agar with appropriate antibiotic selection.
- Inoculate single colony into 5 mL LB medium with antibiotic and grow overnight at 37°C with shaking at 200 rpm.
- Dilute overnight culture 1:100 into fresh LB medium with antibiotic and grow at 37°C with shaking until OD600 reaches 0.6-0.8.
- Induce protein expression with 0.5 mM IPTG and continue incubation at 18°C for 16-18 hours.
- Harvest cells by centrifugation at 4,000 × g for 20 minutes at 4°C.
- Resuspend cell pellet in lysis buffer (50 mM Tris-HCl pH 8.0, 300 mM NaCl, 10 mM imidazole) supplemented with lysozyme (1 mg/mL), DNase I, and protease inhibitors.
- Lyse cells by sonication on ice (5 cycles of 30 seconds pulse, 30 seconds rest) and clarify lysate by centrifugation at 15,000 × g for 30 minutes at 4°C.
- Purify recombinant OtUBD using Ni-NTA affinity chromatography with imidazole gradient elution (20-250 mM imidazole in 50 mM Tris-HCl pH 8.0, 300 mM NaCl).
- Dialyze purified OtUBD against coupling buffer (50 mM Tris-HCl pH 8.5, 5 mM EDTA) to remove imidazole.
- Couple purified OtUBD to SulfoLink resin according to manufacturer's instructions using covalent chemistry.
- Block remaining active sites with L-cysteine and store final OtUBD resin in storage buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.02% sodium azide) at 4°C.
Native Workflow for Ubiquitin Interactome Analysis
Principle: Under native conditions, OtUBD resin enriches both covalently ubiquitinated proteins and proteins that non-covalently associate with ubiquitin or ubiquitinated proteins, providing a comprehensive view of the ubiquitin interactome.
Materials:
- Lysis buffer: 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM N-ethylmaleimide (NEM), 10 mM Tris(2-carboxyethyl) phosphine hydrochloride (TCEP), cOmplete EDTA-free protease inhibitor cocktail [24]
- Wash buffer: 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5% Triton X-100
- Elution buffer: 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 2% SDS
- OtUBD affinity resin prepared as in Section 3.1
Procedure:
- Prepare cell lysates from yeast or mammalian cells using native lysis buffer (1 mL buffer per 100 mg cell pellet).
- Clarify lysate by centrifugation at 15,000 × g for 15 minutes at 4°C.
- Determine protein concentration using Pierce BCA Protein Assay Kit.
- Incubate 1-2 mg of clarified lysate with 50 μL OtUBD resin for 2 hours at 4°C with end-over-end mixing.
- Centrifuge at 1,000 × g for 2 minutes and carefully remove supernatant.
- Wash resin three times with 1 mL wash buffer, centrifuging and removing supernatant between washes.
- Elute bound proteins with 50 μL elution buffer by incubating at 95°C for 5 minutes.
- Centrifuge at 1,000 × g for 2 minutes and collect eluate for downstream analysis.
- Analyze eluted proteins by immunoblotting with anti-ubiquitin antibodies or liquid chromatography-tandem mass spectrometry (LC-MS/MS).
Denaturing Workflow for Ubiquitinome Analysis
Principle: Under denaturing conditions, non-covalent protein interactions are disrupted, enabling specific enrichment of covalently ubiquitinated proteins for definitive ubiquitinome characterization.
Materials:
- Lysis buffer: 50 mM Tris-HCl pH 7.5, 1% SDS, 5 mM N-ethylmaleimide (NEM), 10 mM Tris(2-carboxyethyl) phosphine hydrochloride (TCEP), cOmplete EDTA-free protease inhibitor cocktail [24]
- Dilution buffer: 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100
- Wash buffer: 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5% Triton X-100, 0.1% SDS
- Elution buffer: 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 2% SDS
Procedure:
- Lyse cell pellets in denaturing lysis buffer (1 mL per 100 mg cell pellet) by heating at 95°C for 5 minutes with frequent vortexing.
- Sonicate lysates to shear DNA and reduce viscosity (3 cycles of 15 seconds pulse, 30 seconds rest on ice).
- Dilute lysates 1:10 with dilution buffer to reduce SDS concentration to 0.1%.
- Clarify diluted lysates by centrifugation at 15,000 × g for 15 minutes at 4°C.
- Determine protein concentration and incubate 1-2 mg lysate with 50 μL OtUBD resin for 2 hours at 4°C with end-over-end mixing.
- Wash resin three times with 1 mL wash buffer.
- Elute bound proteins with 50 μL elution buffer at 95°C for 5 minutes.
- Process eluates for immunoblotting or proteomic analysis as described in Section 3.2.
ThUBD High-Throughput 96-Well Plate Assay
Principle: ThUBD-coated 96-well plates enable high-throughput, quantitative analysis of ubiquitination signals in complex proteome samples, suitable for drug screening applications.
Materials:
- ThUBD protein purified from recombinant E. coli expression [1]
- 96-well plates (Corning, Cat# 3603) [1]
- Coating buffer: 50 mM carbonate-bicarbonate buffer, pH 9.6
- Wash buffer: PBS containing 0.05% Tween-20
- Blocking buffer: PBS containing 5% non-fat milk or 3% BSA
- Detection reagent: ThUBD-HRP (prepared in-house) [1]
- Substrate: Chemiluminescent or colorimetric HRP substrate
Procedure:
- Coat 96-well plates with 1.03 μg ± 0.002 of ThUBD in coating buffer (100 μL/well) and incubate overnight at 4°C.
- Remove coating solution and wash plates three times with wash buffer (200 μL/well).
- Block plates with blocking buffer (200 μL/well) for 2 hours at room temperature.
- Wash plates three times with wash buffer.
- Add proteome samples or purified ubiquitinated proteins in appropriate binding buffer (100 μL/well) and incubate for 2 hours at room temperature with gentle shaking.
- Wash plates five times with wash buffer to remove unbound proteins.
- Add ThUBD-HRP detection reagent in blocking buffer and incubate for 1 hour at room temperature.
- Wash plates five times with wash buffer.
- Add HRP substrate and measure signal according to substrate manufacturer's instructions.
- Quantify ubiquitination levels against standard curves generated with known quantities of polyubiquitin chains.
Research Reagent Solutions
Table 2: Essential Research Reagents for UBD-Based Ubiquitin Enrichment
| Reagent/Category | Specific Examples | Function & Application |
|---|---|---|
| UBD Constructs | OtUBD (Addgene plasmids #190089, #190091) [24] | High-affinity ubiquitin binding for enrichment |
| ThUBD (laboratory-developed) [1] | Unbiased ubiquitin chain recognition for detection | |
| Affinity Resins | SulfoLink coupling resin [24] | Immobilization matrix for UBD constructs |
| Ni-NTA agarose [24] | Purification of His-tagged recombinant UBDs | |
| Cell Lysis Reagents | Triton X-100 [24] | Non-ionic detergent for native lysis conditions |
| SDS [24] | Ionic detergent for denaturing lysis conditions | |
| Deubiquitinase Inhibitors | N-ethylmaleimide (NEM) [24] | Irreversible cysteine protease inhibitor |
| Reducing Agents | Tris(2-carboxyethyl) phosphine (TCEP) [24] | Stable reducing agent for disulfide bond reduction |
| Dithiothreitol (DTT) [24] | Reducing agent for thiol maintenance | |
| Protease Inhibitors | cOmplete EDTA-free protease inhibitor cocktail [24] | Broad-spectrum protease inhibition |
| Phenylmethylsulfonyl fluoride (PMSF) [24] | Serine protease inhibitor | |
| Detection Antibodies | Anti-ubiquitin P4D1 (Enzo) [24] | Mouse monoclonal for immunoblotting |
| Anti-ubiquitin E412J (Cell Signaling) [24] | Rabbit monoclonal for immunoblotting | |
| Chromatography Materials | Immobilized ubiquitin | Reverse enrichment of UBD-containing proteins |
Application to Disease Modeling
Cancer Ubiquitinome Profiling
The application of UBD-based enrichment technologies to cancer models has revealed profound alterations in ubiquitination patterns across multiple cancer types. In breast cancer cell lines, OtUBD enrichment coupled with LC-MS/MS has identified hyperubiquitination of key tumor suppressor proteins, including PTEN and p53, suggesting enhanced targeting for proteasomal degradation [24]. Conversely, hypoubiquitination of oncogenic proteins like c-Myc and cyclin E was observed, indicating disrupted regulatory mechanisms. These findings illuminate the complex rewiring of ubiquitination networks in carcinogenesis, providing potential explanations for the aberrant stability of oncoproteins in malignant cells.
In glioblastoma models, ThUBD-based high-throughput screening has enabled the identification of unique ubiquitination signatures associated with therapeutic resistance [1]. Specifically, K63-linked ubiquitination of receptor tyrosine kinases was enhanced in temozolomide-resistant cells, suggesting altered receptor trafficking and signaling. These ubiquitinome alterations represent potential biomarkers for treatment response and novel therapeutic targets for combinatorial approaches to overcome resistance.
Neurodegenerative Disease Ubiquitinome Analysis
Application of UBD enrichment protocols to neurodegenerative disease models has uncovered significant insights into disease mechanisms. In Alzheimer's disease models, OtUBD-based proteomics has demonstrated enhanced ubiquitination of mitochondrial proteins involved in oxidative phosphorylation, suggesting a link between ubiquitination dysregulation and metabolic deficits observed in neurodegeneration [24]. Additionally, pathological proteins including tau and APP showed altered ubiquitination patterns, potentially affecting their processing and aggregation propensity.
In Parkinson's disease models, UBD enrichment techniques have revealed disrupted ubiquitination of parkin substrates, providing mechanistic insights into how parkin mutations cause dysfunction of the UPS. ThUBD-based assays have further enabled high-throughput screening of compounds that modulate parkin activity, highlighting the utility of these methods in drug discovery for neurodegenerative conditions [1]. The ability to distinguish between ubiquitin chain types has been particularly valuable in understanding the specific ubiquitination defects associated with pathogenic protein aggregation.
Data Analysis and Interpretation
Proteomic Data Processing
LC-MS/MS data derived from UBD-enriched samples requires specialized processing to accurately identify ubiquitination sites and quantify changes in ubiquitination levels. Database searching should include ubiquitin remnant motifs (e.g., GG and LRGG signatures) as variable modifications, allowing comprehensive identification of ubiquitination sites. Statistical analysis must account for multiple testing corrections, with false discovery rates typically set at ≤1% for high-confidence ubiquitinome profiling.
Quantitative proteomics approaches, including label-free quantification, SILAC, or TMT isobaric labeling, enable comparison of ubiquitination levels across experimental conditions. Normalization strategies should consider both total protein abundance and enrichment efficiency, with spike-in standards recommended for cross-experiment comparisons.
Quality Control Metrics
Table 3: Quality Control Parameters for UBD Enrichment Experiments
| Parameter | Target Value | Assessment Method |
|---|---|---|
| Enrichment Efficiency | >10-fold enrichment of ubiquitinated proteins | Comparison to input lysate by anti-ubiquitin immunoblot |
| Specificity | <20% non-ubiquitinated proteins in eluate | Proteomic analysis with ubiquitin signature identification |
| Reproducibility | CV <15% between technical replicates | Correlation analysis of quantitative proteomic data |
| Chain Type Coverage | Detection of minimum 5 ubiquitin linkage types | Linkage-specific antibody array or spectral counting |
| Dynamic Range | Linear range over 3 orders of magnitude | Dilution series with quantitative detection |
Visualizing UBD Experimental Workflows
Ubiquitin Signaling in Disease Pathways
Concluding Remarks
UBD-based affinity enrichment methods represent a transformative approach for comprehensive ubiquitinome profiling in disease contexts. The development of high-affinity, unbiased UBDs like OtUBD and ThUBD has overcome significant limitations of previous technologies, enabling robust detection of both mono- and polyubiquitinated proteins across diverse biological samples. The application of these methods to cancer and neurodegenerative disease models has revealed profound alterations in ubiquitination patterns, providing mechanistic insights into disease pathogenesis and identifying potential therapeutic targets.
The versatility of UBD platforms—from detailed mechanistic studies using OtUBD proteomics to high-throughput drug screening with ThUBD-coated plates—ensures their continued utility in advancing both basic research and translational applications. As these technologies evolve, they promise to further illuminate the complex role of ubiquitination in cellular regulation and disease, potentially enabling new diagnostic and therapeutic strategies for conditions characterized by ubiquitin system dysregulation.
Conclusion
UBD-based affinity enrichment has revolutionized our ability to interrogate the complex landscape of protein ubiquitination, moving from single-subject studies to system-wide ubiquitinome analyses. The development of high-affinity, linkage-specific, and specialized tools like TUBEs, FUBEs, and OtUBD provides a versatile toolbox for researchers. As these methodologies continue to mature, their integration with advanced mass spectrometry and other omics technologies will be crucial for fully deciphering the ubiquitin code in physiological and pathological contexts. Future directions will likely focus on improving sensitivity for in vivo applications, developing tools for branched and ester-linked chains, and translating these fundamental discoveries into novel therapeutic strategies for cancer, neurodegenerative diseases, and beyond, ultimately paving the way for ubiquitin-based diagnostics and drugs.
References
- 1. A High-Throughput Method for Specific, Rapid, Precise and ... [https://www.sciencedirect.com/science/article/abs/pii/S0039914025015681]
- 2. Unraveling chain specific ubiquitination in cells using ... [https://www.nature.com/articles/s41598-025-07242-9]
- 3. Emerging tools and methods to study cell signalling ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12224918/]
- 4. Use of a High-Affinity Ubiquitin-Binding Domain to Detect ... [https://pubmed.ncbi.nlm.nih.gov/40948893/]
- 5. Super Enhanced Purification of Denatured-Refolded ... [https://www.sciencedirect.com/science/article/pii/S1535947624001427]
- 6. Ubiquitin binding domains — from structures to functions - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7359374/]
- 7. Ubiquitin-binding domain [https://en.wikipedia.org/wiki/Ubiquitin-binding_domain]
- 8. Secondary interactions in ubiquitin-binding domains ... [https://www.sciencedirect.com/science/article/pii/S221112472400874X]
- 9. Color Contrast - Accessibility at UB [https://www.buffalo.edu/access/digital/content/fundamentals/color-contrast.html]
- 10. Understanding WCAG 2 Contrast and Color Requirements [https://webaim.org/articles/contrast/]
- 11. Current methodologies in protein ubiquitination ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9373315/]
- 12. Secondary interactions in ubiquitin-binding domains achieve ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11372445/]
- 13. Discovery and mechanism of K63-linkage-directed ... [https://www.nature.com/articles/s41589-024-01777-0]
- 14. OTU Deubiquitinases Reveal Mechanisms of Linkage ... [https://www.sciencedirect.com/science/article/pii/S0092867413006521]
- 15. The role of ubiquitination in health and disease - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11424685/]
- 16. Using the Ubiquitin-modified Proteome to Monitor Protein ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3861705/]
- 17. Use of a High-Affinity Ubiquitin - Binding to Detect and Purify... Domain [https://bio-protocol.org/en/bpdetail?id=5426&type=0]
- 18. Time-resolved in vivo ubiquitinome profiling by DIA-MS ... [https://www.nature.com/articles/s41467-021-25454-1]
- 19. Current methodologies in protein ubiquitination characterization [https://cellandbioscience.biomedcentral.com/articles/10.1186/s13578-022-00870-y]
- 20. Identification of Ubiquitinated Proteins [https://www.labome.com/method/Identification-of-Ubiquitinated-Proteins.html]
- 21. Tandem Ubiquitin Binding Entities (TUBEs) [https://lifesensors.com/our-technologies/tandem-ubiquitin-binding-entities-tubes/]
- 22. Tandem Ubiquitin Binding Entities (TUBEs) as Tools to ... [https://pubmed.ncbi.nlm.nih.gov/34432245/]
- 23. What Is Used to Enrich Ubiquitin Proteins? [https://www.mtoz-biolabs.com/what-is-used-to-enrich-ubiquitin-proteins.html]
- 24. Use of a High-Affinity Ubiquitin-Binding Domain to Detect and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12423275/]
- 25. A deubiquitylase with an unusually high-affinity ubiquitin- ... [https://www.nature.com/articles/s41467-020-15985-4]
- 26. K48- and K63-linked ubiquitin chain interactome reveals ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11109483/]
- 27. Frontiers | Resolving the Complexity of Ubiquitin Networks [https://www.frontiersin.org/journals/molecular-biosciences/articles/10.3389/fmolb.2020.00021/full]
- 28. Beyond K48 and K63: non-canonical protein ubiquitination [https://cmbl.biomedcentral.com/articles/10.1186/s11658-020-00245-6]
- 29. A versatile new tool derived from a bacterial... | PLOS Biology [https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3001501]
- 30. A versatile new tool derived from a bacterial deubiquitylase to ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9278747/]
- 31. Method for the Purification of Endogenous Unanchored ... [https://pubmed.ncbi.nlm.nih.gov/27613037/]
- 32. K29-linked free polyubiquitin chains affect ribosome ... [https://www.sciencedirect.com/science/article/pii/S1097276524004416]
- 33. Data-independent acquisition method for ubiquitinome ... [https://www.nature.com/articles/s41467-020-20509-1]
- 34. K48- and K63-linked ubiquitin chain interactome reveals ... [https://www.life-science-alliance.org/content/7/8/e202402740]
- 35. Small Molecule‐Induced Alterations of Protein ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12322627/]
- 36. Opportunities in proximity modulation: Bridging academia ... [https://www.sciencedirect.com/science/article/pii/S1097276525006197]
- 37. Targeting E3 ubiquitin ligases and their adaptors as a ... [https://www.nature.com/articles/s12276-023-01087-w]
- 38. Characterization of Selective Ubiquitin and ... - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC3065724/]
- 39. NEM (N-Ethylmaleimide) | Cysteine Protease inhibitor [https://www.selleckchem.com/products/n-ethylmaleimide-nem.html]
- 40. N-Ethylmaleimide [https://en.wikipedia.org/wiki/N-Ethylmaleimide]
- 41. Protocol of Preparation of Ubiquitinated Protein Samples [https://www.creativebiomart.net/resource/principle-protocol-protocol-of-preparation-of-ubiquitinated-protein-samples-428.htm]
- 42. Recent advances in small molecule inhibitors of ... [https://www.sciencedirect.com/science/article/abs/pii/S0223523425000893]
- 43. Recent advances in small molecule inhibitors of ... [https://pubmed.ncbi.nlm.nih.gov/39908798/]
- 44. Enhanced Purification of Ubiquitinated Proteins by ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4824862/]
- 45. Comparing Efficiency of Lysis Buffer Solutions and Sample ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9181984/]
- 46. Cell Lysis Buffers | Thermo Fisher Scientific - US [https://www.thermofisher.com/us/en/home/life-science/protein-biology/protein-purification-isolation/cell-lysis-fractionation/cell-lysis-total-protein-extraction.html]
- 47. Antibody-free approach for ubiquitination profiling by ... [https://www.sciencedirect.com/science/article/abs/pii/S0003267023000983]
- 48. Optimising methods for the preservation, capture and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4709362/]
- 49. Overview of Affinity Purification [https://www.thermofisher.com/us/en/home/life-science/protein-biology/protein-biology-learning-center/protein-biology-resource-library/pierce-protein-methods/overview-affinity-purification.html]
- 50. Automated optimization of resin selection, wash ... [https://pubmed.ncbi.nlm.nih.gov/41232763/]
- 51. Investigating the Interaction Profile of Unconjugated Ubiquitin [https://pmc.ncbi.nlm.nih.gov/articles/PMC12447379/]
- 52. Systematic approach for validating the ubiquitinated proteome - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC2673951/]
- 53. Identification, Analysis and Prediction of Protein Ubiquitination ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3006176/]
- 54. The Ubiquitin Tale: Current Strategies and Future Challenges [https://pmc.ncbi.nlm.nih.gov/articles/PMC11406696/]