TUBE Technology: A Comprehensive Guide to Isolating and Studying Ubiquitinated Proteins
This article provides a detailed overview of Tandem Ubiquitin-Binding Entities (TUBEs), powerful molecular tools designed to overcome long-standing challenges in ubiquitin research.
TUBE Technology: A Comprehensive Guide to Isolating and Studying Ubiquitinated Proteins
Abstract
This article provides a detailed overview of Tandem Ubiquitin-Binding Entities (TUBEs), powerful molecular tools designed to overcome long-standing challenges in ubiquitin research. Aimed at researchers, scientists, and drug development professionals, we cover the foundational science behind TUBEs, including their design based on ubiquitin-associated (UBA) domains and their nanomolar affinity for polyubiquitin chains. The scope extends to practical, step-by-step methodological applications for the isolation and enrichment of ubiquitinated proteins from native cell and tissue extracts, even in the absence of standard protease inhibitors. We further address troubleshooting and optimization strategies to enhance experimental outcomes and include a critical validation and comparative analysis of TUBEs against other techniques, such as diGly antibody enrichment. The content synthesizes the latest research to demonstrate how TUBEs are revolutionizing the decoding of the ubiquitin code in biomedical and clinical research.
Decoding the Ubiquitin Code: The Science and Design of TUBEs
Protein ubiquitination is a versatile and reversible post-translational modification (PTM) that regulates virtually all aspects of eukaryotic biology, including proteasomal degradation, cell signaling, DNA repair, and immune responses [1] [2]. This complexity arises from the ability of ubiquitin to form diverse polymeric chains through eight different linkage types (Met1 and Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, Lys63), creating a sophisticated "ubiquitin code" that dictates specific cellular outcomes [3] [2]. The reversibility of this process, mediated by deubiquitinases (DUBs), along with the low stoichiometry of endogenous ubiquitination and the lability of ubiquitin conjugates, presents significant methodological challenges for researchers [4] [5].
Table 1: Key Challenges in Ubiquitin Research
| Challenge | Impact on Research |
|---|---|
| Reversibility by DUBs | Rapid deubiquitination during lysis and processing, leading to loss of signal [4] |
| Low Stoichiometry | Difficulty detecting endogenous ubiquitination events amidst non-modified proteins [4] |
| Structural Complexity | Difficulty distinguishing between chain linkage types and architectures [3] [4] |
| Substrate Lability | Proteasomal degradation of ubiquitinated proteins before they can be analyzed [6] |
| PTM Crosstalk | Interdependence with phosphorylation, acetylation, etc., complicating analysis [7] [8] |
The ubiquitin network's complexity is managed in cells by the orchestrated interplay of hundreds of enzymes, including 2 E1 activating enzymes, approximately 40 E2 conjugating enzymes, over 600 E3 ligases, and around 100 DUBs encoded by the human genome [4] [2]. For researchers, capturing this intricate and dynamic system requires tools that can not only isolate ubiquitinated proteins with high affinity but also preserve their often transient and unstable state throughout the experimental process [6] [5].
Figure 1: The Complexity of the Ubiquitin Code. Ubiquitin can generate diverse signals through monoubiquitination, homotypic polyubiquitin chains, and complex atypical chains (mixed/branched). These ubiquitin modifications are further complicated by crosstalk with other post-translational modifications (PTMs).
TUBEs: A Strategic Tool for Isolating Ubiquitinated Proteins
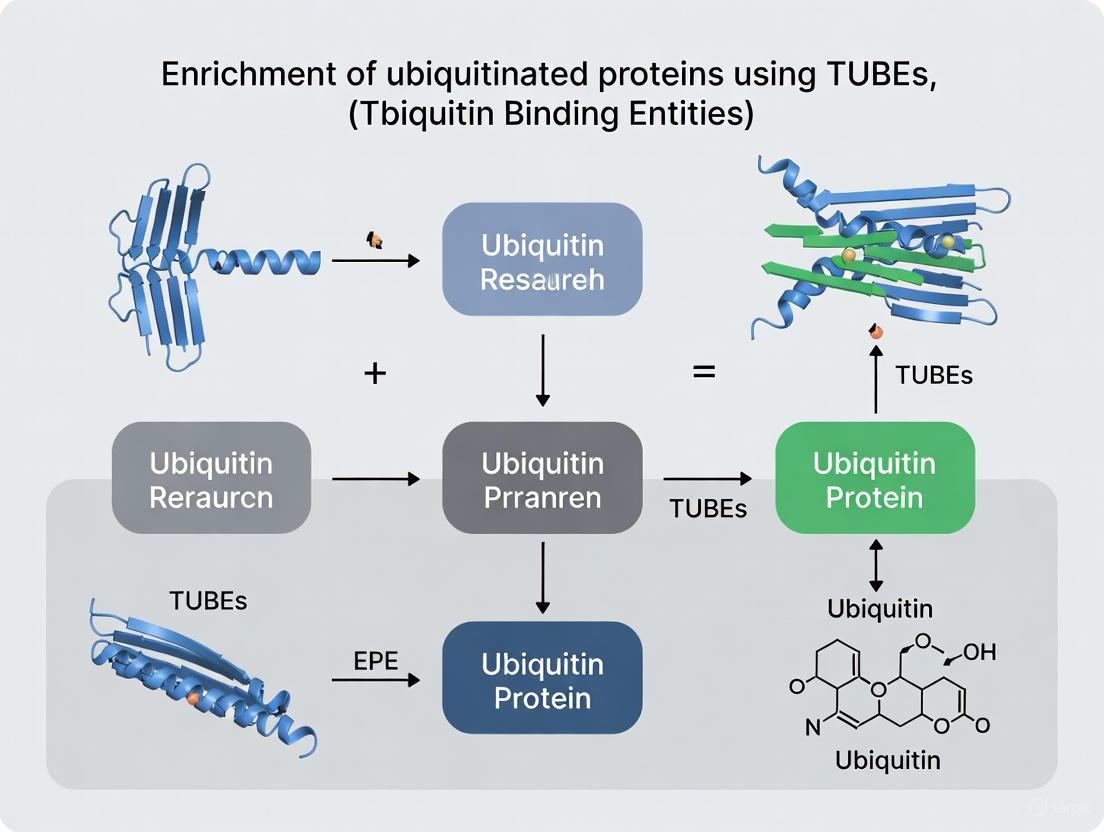
Tandem Ubiquitin Binding Entities (TUBEs) are engineered protein reagents containing multiple ubiquitin-binding domains (UBDs) arranged in tandem, which confer nanomolar affinity (Kd 1-10 nM) for polyubiquitin chains [6] [5]. This multi-valent design allows TUBEs to outperform single UBDs and traditional ubiquitin antibodies by overcoming the inherently weak affinity of individual ubiquitin-binding interactions [4] [9]. The strategic arrangement of UBDs creates a synergistic effect, significantly increasing avidity for polyubiquitin chains and enabling efficient capture of ubiquitinated proteins from complex lysates without the need for epitope-tagged ubiquitin overexpression [6] [5].
A critical functional advantage of TUBEs is their ability to protect the ubiquitin signal they bind. By physically shielding polyubiquitin chains from the action of DUBs and hindering access by the proteasome, TUBEs prevent the deubiquitination and degradation of target proteins that would otherwise occur during cell lysis and sample processing, even in the absence of standard protease and DUB inhibitors [6]. This preservation of labile ubiquitin modifications provides a more accurate snapshot of the cellular ubiquitination state.
Research Reagent Solutions
Table 2: Essential Research Reagents for TUBE-Based Ubiquitin Studies
| Reagent | Function & Application | Key Feature |
|---|---|---|
| Pan-Selective TUBEs | General capture of all polyubiquitin chains; ideal for global ubiquitylome studies [6]. | Binds all linkage types with high affinity (Kd ~1-10 nM). |
| Linkage-Selective TUBEs (K48, K63, M1) | Isolate specific chain types to study linkage-specific functions [6] [5]. | Elucidates consequences of specific ubiquitin codes. |
| TAMRA-Labeled TUBEs (e.g., UM202) | Imaging ubiquitin dynamics in live or fixed cells [6]. | Fluorophore on fusion tag doesn't interfere with ubiquitin binding. |
| Agarose-Conjugated TUBEs (e.g., UM501M) | Affinity pulldown of ubiquitinated proteins for MS or WB [6]. | Compatible with mass spectrometry and western blotting. |
| Microtiter Plate-Immobilized TUBEs | High-throughput screening assays for drug discovery [6] [5]. | Enables screening for ubiquitination modulators. |
Detailed Experimental Protocols
Protocol 1: Affinity Purification of Ubiquitinated Proteins Using TUBEs
This protocol describes the isolation of polyubiquitinated proteins from mammalian cell lysates using agarose-conjugated TUBEs, suitable for subsequent analysis by western blotting or mass spectrometry.
Reagents and Equipment:
- Agarose-conjugated Pan-TUBE (e.g., LifeSensors UM501M)
- Lysis Buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, supplemented with protease inhibitors (optional when using TUBEs)
- Wash Buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% NP-40
- Elution Buffer: 100 mM Tris-HCl (pH 6.8), 4% SDS, 20% glycerol, 0.02% bromophenol blue (for direct western loading) OR 2x Laemmli buffer
- Control: Beads conjugated with an irrelevant protein
- Cell scrappers, microcentrifuge, rotator
Procedure:
- Cell Lysis: Harvest cells by scraping and lyse in ice-cold Lysis Buffer. For a 10 cm plate, use 500 μL to 1 mL of buffer. Incubate on ice for 10 minutes.
- Clarification: Centrifuge the lysate at 17,000 x g for 15 minutes at 4°C. Transfer the supernatant to a new tube. Determine protein concentration.
- Binding: Incubate 500 μg - 2 mg of total protein lysate with 20 μL of settled Pan-TUBE agarose beads. Adjust the final volume to 500 μL with Lysis Buffer if necessary.
- Capture: Rotate the mixture for 2 hours at 4°C to maximize binding.
- Washing: Centrifuge samples at 2,500 x g for 2 minutes to pellet beads. Carefully remove the supernatant.
- Wash the beads three times with 500 μL of Wash Buffer, rotating for 5 minutes per wash.
- Perform a final quick wash with 50 mM Tris-HCl (pH 7.5) to remove detergent.
- Elution:
- For Western Blotting: Add 40 μL of Elution Buffer or 2x Laemmli buffer to the beads. Heat at 95°C for 5-10 minutes. Centrifuge and load the supernatant onto a gel.
- For Mass Spectrometry: Use a milder, MS-compatible elution such as 8 M urea or low-pH buffer. Alternatively, on-bead trypsin digestion can be performed.
Technical Notes:
- TUBEs protect from DUBs, but for maximum preservation, work quickly and keep samples cold.
- Include a control with non-TUBE beads to identify non-specifically bound proteins.
- For mass spectrometry, consider crosslinking the TUBEs to the beads to prevent leakage of the affinity reagent.
Protocol 2: Detection of Polyubiquitinated Proteins by TUBE-Based Immunoblotting
This method uses TUBEs as a replacement for traditional ubiquitin antibodies in western blotting, offering enhanced sensitivity for detecting polyubiquitin chains.
Reagents and Equipment:
- TAMRA-TUBE (e.g., LifeSensors UM202) or other labeled TUBE
- Standard Western Blotting equipment and reagents
- Blocking Buffer: 5% non-fat dry milk in TBST (Tris-Buffered Saline with 0.1% Tween-20)
- Primary Antibody against your protein of interest
- Fluorescence-compatible imaging system (if using TAMRA-TUBE)
Procedure:
- Electrophoresis and Transfer: Separate proteins by SDS-PAGE and transfer to a PVDF membrane using standard protocols.
- Blocking: Block the membrane with Blocking Buffer for 1 hour at room temperature.
- Probing with TUBE: Dilute the TAMRA-TUBE in Blocking Buffer (e.g., 1:1000). Incubate the membrane with the TUBE solution for 2 hours at room temperature or overnight at 4°C.
- Washing: Wash the membrane three times for 10 minutes each with TBST.
- Imaging: If using TAMRA-TUBE, directly image the membrane using a fluorescence scanner with the appropriate channel (Ex. = 540 nm, Emm. = 578 nm).
- Reprobing (Optional): After imaging, the same membrane can be stripped and re-probed with a primary antibody against your protein of interest to confirm the identity of the ubiquitinated species.
Technical Notes:
- This method specifically detects polyubiquitinated proteins due to the multi-valent binding requirement of TUBEs.
- Direct fluorescence detection avoids secondary antibody cross-reactivity and offers a cleaner signal.
- The reprobing step confirms that the high-molecular-weight smears correspond to the protein of interest.
Figure 2: TUBE-Based Affinity Purification Workflow. Cellular lysate is incubated with TUBE-conjugated beads, which selectively bind and protect polyubiquitinated proteins from deubiquitinating enzymes (DUBs) and proteasomal degradation during isolation.
Applications in Drug Discovery and Targeted Protein Degradation
TUBE technology has found significant utility in the burgeoning field of targeted protein degradation (TPD), particularly in the development and validation of PROTACs (Proteolysis-Targeting Chimeras) and molecular glues [6] [5]. These bifunctional molecules recruit an E3 ubiquitin ligase to a protein of interest (POI), inducing its polyubiquitination and subsequent proteasomal degradation. TUBEs provide a direct means to monitor the efficiency of this process by enabling researchers to confirm and quantify the polyubiquitination of the POI in response to PROTAC treatment [5].
The implementation of TUBEs in high-throughput screening (HTS) platforms, where they are immobilized on microtiter plates, allows for the rapid assessment of polyubiquitination levels in vitro or in cellular models [6]. This application accelerates the identification of effective degraders and the establishment of structure-activity relationships, which are crucial steps in the drug discovery pipeline. By offering a more specific and sensitive readout of the key molecular event—ubiquitination—TUBEs help distinguish true mechanistic hits from false positives that may merely reduce target protein levels through indirect mechanisms, such as transcriptional repression.
Table 3: TUBE Applications in Research and Drug Discovery
| Application Field | Specific Use Case | Benefit |
|---|---|---|
| Basic Research | Global ubiquitylome analysis via mass spectrometry [4]. | Identifies novel substrates and sites without genetic manipulation. |
| Disease Mechanism | Studying ubiquitination in patient tissue samples [4] [2]. | Works in clinically relevant, non-engineered samples. |
| Biomarker Discovery | Identifying ubiquitination signatures in cancer or neurodegeneration [6] [2]. | High-affinity capture enables detection of low-abundance modifications. |
| PROTAC/TPD Development | Confirming target ubiquitination in cellular and in vitro assays [6] [5]. | Directly measures the key pharmacological event induced by degraders. |
| High-Throughput Screening | Plate-based assays to find ubiquitination modulators [6]. | Enables rapid screening of compound libraries. |
The study of the ubiquitin system demands sophisticated tools that can address the inherent challenges of low stoichiometry, structural complexity, and dynamic reversibility. TUBE technology, with its high affinity, linkage selectivity, and protective functionality, provides a robust methodological foundation for isolating and preserving polyubiquitinated proteins. The detailed protocols outlined herein for affinity purification, blotting, and drug discovery applications empower researchers to decipher the complex ubiquitin code with greater accuracy and reliability, ultimately advancing both basic biological understanding and the development of novel therapeutics targeting the ubiquitin-proteasome system.
Ubiquitin-binding domains (UBDs) are modular protein elements that bind non-covalently to the protein modifier ubiquitin, serving as critical interpreters of the ubiquitin code within eukaryotic cells [10]. These specialized domains facilitate the recognition of ubiquitinated proteins and transduce ubiquitin signals into diverse cellular outcomes, including protein degradation, DNA repair, immune signaling, and endocytic trafficking [10] [11]. The versatility of ubiquitin signaling originates from the diversity of ubiquitin modifications—ranging from single ubiquitin molecules (monoubiquitination) to complex polyubiquitin chains connected through different linkage types—each capable of being recognized by specific UBDs [10] [4].
The significance of UBDs extends throughout nearly all cellular processes, with dysregulation in ubiquitin-UBD interactions contributing to pathologies such as cancer, neurodegenerative diseases, and inflammatory disorders [10] [4]. As of current research, more than 20 distinct families of UBDs have been identified, encompassing a remarkable variety of structural folds that all enable specific recognition of ubiquitin surfaces [10] [11]. This article explores the structural and functional diversity of UBDs and provides detailed protocols for their application in enriching ubiquitinated proteins, with particular focus on Tandem Ubiquitin Binding Entities (TUBEs) and related technologies that have revolutionized the study of ubiquitin signaling.
The Structural and Functional Diversity of UBDs
Structural Classification and Ubiquitin Recognition Mechanisms
UBDs exhibit remarkable structural diversity while maintaining the common function of ubiquitin recognition. These domains can be structurally classified into several major categories: α-helical domains, zinc fingers, pleckstrin homology (PH) domains, Ubc-like domains, and other distinct folds [10]. Despite their structural differences, most UBDs interact with a common hydrophobic patch on ubiquitin centered around Ile44, though they approach this surface with different binding modes and affinities [10] [11].
α-helical UBDs represent the largest class and include the UBA (Ubiquitin-Associated), UIM (Ubiquitin-Interacting Motif), MIU (Motif Interacting with Ubiquitin), DUIM (Double-sided UIM), CUE (Coupling of Ubiquitin conjugation to ER degradation), and GAT (GGA and TOM) domains [11]. These domains share a common three-helical bundle architecture but employ different binding mechanisms. For instance, the UIM consists of a single α-helix that binds in a shallow hydrophobic groove on ubiquitin, with a conserved alanine residue packing against Ile44 of ubiquitin [11]. In contrast, CUE domains form dimeric structures that enhance ubiquitin binding avidity [11].
Zinc finger UBDs constitute another major class and include the NZF (Npl4 Zinc Finger), A20 ZnF, UBZ (Ubiquitin-Binding Zinc finger), and ZnF UBP domains [10]. These domains utilize zinc coordination to maintain their structural integrity while providing versatile ubiquitin-binding surfaces. The ZnF UBP domain of Isopeptidase T exhibits particularly high affinity for ubiquitin, with a Kd of 2.8 μM [11].
Other UBD classes include the UEV (Ubiquitin-Conjugating Enzyme E2 Variant), Ubc (Ubiquitin-Conjugating enzyme), PRU (Pleckstrin Homology Receptor for Ubiquitin), and GLUE (GRAM-Like Ubiquitin Binding in EAP45) domains [10]. Each class possesses distinct structural features that dictate its specificity for different ubiquitin signals.
Table 1: Major Classes of Ubiquitin-Binding Domains and Their Characteristics
| Structural Class | UBD Types | Representative Proteins | Primary Cellular Functions | Typical Affinity Range (Kd) |
|---|---|---|---|---|
| α-helical | UIM, MIU, DUIM | S5a/Rpn10, Vps27, STAM, EPSINs, RAP80 | Proteasomal degradation, endocytosis, MVB biogenesis, DNA repair | 100 μM - 2 mM (UIM) [11] |
| α-helical | UBA, CUE | Rad23/HR23A, Dsk2, Vps9, TAB2 | Proteasome targeting, kinase regulation, endocytosis | 14-400 μM (UBA) [11] |
| Zinc finger | NZF, A20 ZnF, UBZ, ZnF UBP | NPL4, Vps36, RABEX-5, POL-h, POL-k, IsoT | ERAD, MVB biogenesis, DNA damage tolerance, NF-κB signaling | 2.8-500 μM [10] [11] |
| PH domain | PRU, GLUE | RPN13, EAP45 | Proteasome function, MVB biogenesis | Variable |
| Ubc-like | UEV, Ubc | Uev1/Mms2, UBCH5C | DNA repair, MVB biogenesis, ubiquitin transfer | ~300-510 μM [11] |
Linkage Specificity and Avidity Mechanisms
Different UBDs exhibit distinct preferences for specific ubiquitin chain linkages, which forms the molecular basis for their functional specificity in cellular pathways [10]. For example, the UBA domains of hHR23A and Mud1 selectively bind to K48-linked ubiquitin chains, while the NZF domain of TAK1-binding protein 2 (TAB2) prefers K63-linked chains, and the UBAN domain of NEMO specifically recognizes linear (M1-linked) ubiquitin chains [12]. This linkage specificity enables precise decoding of ubiquitin signals to direct appropriate cellular responses.
Individual UBDs typically bind mono-ubiquitin with weak affinities (Kd > 100 μM), which seems counterintuitive for specific signaling functions [11]. However, cells employ several avidity-enhancing mechanisms to achieve physiologically relevant high-affinity interactions:
- Ubiquitin polymerization: Multiple ubiquitin molecules in chains provide additional binding sites [10]
- Tandem UBD arrangements: Multiple UBDs within a single protein cooperate to bind ubiquitin chains [11]
- Oligomerization of UBD-containing proteins: Increases local UBD concentration [11]
- Cooperative binding with other domains: Simultaneous interaction with ubiquitin and other molecules (e.g., phospholipids) [11]
These avidity mechanisms explain how weak individual interactions can yield highly specific and functional ubiquitin signaling outcomes in cellular environments.
Tandem Ubiquitin Binding Entities (TUBEs) represent a technological breakthrough in the study of protein ubiquitination. TUBEs are engineered recombinant proteins that incorporate multiple UBDs in tandem within a single polypeptide, resulting in dramatically enhanced affinity for polyubiquitin chains through avidity effects [6]. These tools have nanomolar affinities for polyubiquitin chains, overcoming the limitation of weak binding exhibited by individual natural UBDs [6] [13].
The primary advantages of TUBE technology include:
- Enhanced Affinity: TUBEs bind polyubiquitin with Kd values in the low nanomolar range (1-10 nM), approximately 1000-fold higher than individual UBDs [6]
- Protection from Deubiquitination: TUBEs shield ubiquitin chains from deubiquitinating enzymes (DUBs), preserving ubiquitination signals during experimental procedures [6]
- Protection from Proteasomal Degradation: Similarly, TUBEs protect ubiquitinated proteins from proteasomal degradation [6]
- Linkage Specificity Options: Pan-selective TUBEs recognize all ubiquitin chain types, while chain-specific TUBEs (K48-, K63-, or M1-specific) enable precise analysis of particular chain linkages [6] [13]
- Application Versatility: TUBEs can be adapted for various applications including immunoblotting, proteomics, imaging, and high-throughput screening [6] [13]
TUBEs have been particularly valuable in the emerging field of targeted protein degradation, where they facilitate the assessment of PROTAC (Proteolysis Targeting Chimera) and molecular glue efficiency by monitoring target protein ubiquitination [6] [13].
Table 2: Research Reagent Solutions for Ubiquitin Research
| Reagent / Tool | Type | Key Features / Applications | Examples / Sources |
|---|---|---|---|
| Pan-TUBEs | Tandem UBD reagent | Binds all polyubiquitin chain types; protects from DUBs/proteasome; for general ubiquitination detection | LifeSensors UM401M [13] |
| Linkage-Specific TUBEs | Specialized TUBEs | Selective for specific linkages (K48, K63, M1); for studying chain-specific functions | LifeSensors K48 HF, K63, M1 TUBEs [6] [13] |
| OtUBD | High-affinity single UBD | Low nanomolar affinity for ubiquitin; effective for mono- and polyubiquitin enrichment | Recombinantly expressed from O. tsutsugamushi [14] |
| ThUBD | Engineered tandem hybrid UBD | Unbiased recognition of all chain types; higher affinity than natural UBDs | ThUDA20, ThUDQ2 constructs [12] |
| TUBE-Coated Plates | HTS format | 96-well plates coated with TUBEs for high-throughput ubiquitination screening | PROTAC Assay Plates [15] [13] |
| TAMRA-TUBE | Fluorescent TUBE | Fluorescently labeled for imaging applications; monitors intracellular ubiquitination | LifeSensors UM202 [6] |
Detailed Experimental Protocols
Protocol 1: Enrichment of Ubiquitinated Proteins Using OtUBD Affinity Resin
This protocol describes the enrichment of ubiquitinated proteins from baker's yeast and mammalian cell lysates using the high-affinity ubiquitin-binding domain OtUBD derived from Orientia tsutsugamushi [14]. OtUBD exhibits low nanomolar affinity for ubiquitin and can enrich both mono- and poly-ubiquitinated proteins.
Reagents and Equipment
- Plasmids: pRT498-OtUBD (Addgene #190089) or pET21a-cys-His6-OtUBD (Addgene #190091) [14]
- Cell Lysis Buffer: 50 mM Na₂HPO₄ (pH 8.0), 500 mM NaCl, 0.01% SDS, 5% glycerol, supplemented with complete EDTA-free protease inhibitor cocktail and 1 mM N-ethylmaleimide (NEM) [14]
- Wash Buffer: 50 mM NH₄HCO₃ with 5 mM iodoacetamide, followed by 50 mM NH₄HCO₃ without iodoacetamide [14]
- Elution Buffer: 1× SDS-PAGE loading buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 0.1% bromophenol blue, 1% β-mercaptoethanol) [14]
- Chromatography Resin: SulfoLink Coupling Resin or similar thiol-reactive support [14]
- Additional Equipment: Standard cell culture equipment, sonicator, centrifuge capable of 70,000 × g, glass beads (for yeast), and incubation rotator [14]
Step-by-Step Procedure
OtUBD Purification and Immobilization:
- Express recombinant OtUBD in E. coli BL21(DE3) using 0.5 mM IPTG induction for 4 hours at 30°C [14]
- Purify the protein using glutathione-sepharose (for GST-tagged constructs) or Ni-NTA agarose (for His-tagged constructs) [14]
- Immobilize purified OtUBD on SulfoLink coupling resin according to manufacturer's instructions [14]
- Store the prepared OtUBD resin at 4°C in PBS with 30% glycerol for stability [14]
Cell Lysis and Sample Preparation:
- For yeast cells: Harvest cells during early log phase (OD₆₀₀ = 1.0) and lyse using glass beads in lysis buffer [14]
- For mammalian cells: Culture to confluence, harvest, and lyse by sonication in lysis buffer [14]
- Clarify lysates by centrifugation at 70,000 × g for 30 minutes at 4°C [14]
- Determine protein concentration using Bradford or BCA assay [14]
Affinity Enrichment:
- Incubate clarified cell lysate (1-10 mg total protein) with OtUBD resin (50-100 μL bed volume) for 30 minutes at 4°C with constant rotation [14]
- Wash the resin sequentially with: (a) lysis buffer, (b) wash buffer with iodoacetamide, and (c) wash buffer without iodoacetamide [14]
- Elute bound ubiquitinated proteins by boiling in 1× SDS-PAGE loading buffer for 5-10 minutes [14]
Downstream Analysis:
Diagram 1: OtUBD Affinity Enrichment Workflow
Protocol 2: TUBE-Based Pull-Down for Linkage-Specific Ubiquitination Analysis
This protocol utilizes chain-specific TUBEs to selectively capture proteins modified with specific ubiquitin chain linkages, enabling analysis of context-dependent ubiquitination events [13].
Reagents and Equipment
- TUBE Reagents: Pan-selective TUBEs (LifeSensors UM401M) and chain-specific TUBEs (K48-, K63-, or M1-specific) [6] [13]
- Cell Lysis Buffer: Optimized to preserve polyubiquitination, typically containing protease inhibitors, DUB inhibitors (NEM or iodoacetamide), and proteasome inhibitors (MG132) if studying degradation targets [13]
- Magnetic Beads: TUBE-conjugated magnetic beads (e.g., LifeSensors UM401M) for easy manipulation [13]
- Wash Buffers: Standard physiological buffer (e.g., PBS or Tris-buffered saline) with mild detergent (0.1% Tween-20) [13]
- Elution Buffer: 2× Laemmli buffer or specific competing agents (e.g., free ubiquitin) [13]
Step-by-Step Procedure
Cell Treatment and Lysis:
- Treat cells with appropriate stimuli (e.g., L18-MDP for K63 ubiquitination of RIPK2 or PROTACs for K48 ubiquitination) for specified durations [13]
- Lyse cells in optimized lysis buffer (500 μL per 10⁷ cells) with constant agitation at 4°C for 30 minutes [13]
- Clarify lysates by centrifugation at 15,000 × g for 15 minutes at 4°C [13]
- Determine protein concentration and adjust samples to equal concentrations [13]
TUBE Pull-Down:
- Incubate equal protein amounts (200-500 μg) with 25 μL of TUBE-conjugated magnetic beads for 2 hours at 4°C with rotation [13]
- For comparative studies, use parallel pull-downs with different chain-specific TUBEs (pan-, K48-, K63-specific) [13]
- Collect beads using a magnetic separator and carefully remove supernatant [13]
Washing and Elution:
Analysis of Captured Proteins:
Diagram 2: Chain-Specific TUBE Analysis Workflow
Protocol 3: ThUBD-Based High-Throughput Ubiquitination Detection
This protocol utilizes engineered Tandem hybrid UBDs (ThUBDs) coated on 96-well plates for high-throughput, sensitive detection of ubiquitinated proteins with minimal linkage bias [15] [12].
Reagents and Equipment
- ThUBD-Coated Plates: Corning 3603-type 96-well plates coated with 1.03 μg/well of ThUBD [15]
- Blocking Buffer: 3% BSA in TBST (Tris-buffered saline with 0.1% Tween-20) [15]
- Binding Buffer: PBS (pH 7.4) with 0.1% BSA and 0.02% Tween-20 [15]
- Detection Reagent: ThUBD-HRP conjugate (in-house prepared) [15]
- Wash Buffer: TBST (25 mM Tris, 150 mM NaCl, 0.1% Tween-20, pH 7.4) [15]
- Substrate Solution: Chemiluminescent HRP substrate suitable for plate readers [15]
Step-by-Step Procedure
Plate Preparation:
Sample Binding:
Detection:
- Wash plates five times with wash buffer [15]
- Add 100 μL/well of ThUBD-HRP detection reagent (diluted 1:1000 in binding buffer) and incubate for 1 hour at room temperature [15]
- Wash plates five times with wash buffer [15]
- Add 100 μL/well of chemiluminescent substrate and measure signal using a plate reader [15]
Data Analysis:
Applications in Drug Discovery and PROTAC Development
TUBE-based technologies have become indispensable tools in modern drug discovery, particularly in the development and characterization of PROTACs (Proteolysis Targeting Chimeras) and molecular glues [6] [13]. These bifunctional molecules redirect E3 ubiquitin ligases to target specific proteins of interest for ubiquitination and degradation, representing a promising therapeutic strategy for previously "undruggable" targets [13].
The application of chain-specific TUBEs enables researchers to:
- Verify successful target ubiquitination by PROTACs [13]
- Determine the linkage specificity of PROTAC-induced ubiquitination (typically K48-linked for degradation) [13]
- Monitor time-dependent changes in ubiquitination status [13]
- Assess efficiency of different PROTAC designs in high-throughput formats [13]
Table 3: Quantitative Performance Comparison of UBD Technologies
| Technology | Affinity for Polyubiquitin | Chain Linkage Bias | Detection Sensitivity | Throughput Capacity |
|---|---|---|---|---|
| Single UBDs | Low (Kd: 2-500 μM) [11] | Variable; often linkage-specific [12] | Low to moderate | Low |
| TUBEs | High (Kd: 1-10 nM) [6] | Pan-selective or chain-specific options available [6] | High | Medium (solution-based) |
| OtUBD | High (low nM range) [14] | Minimal bias; works on mono- and polyubiquitin [14] | High | Medium (solution-based) |
| ThUBD | Very high (enhanced affinity) [12] | Minimal bias across 7 lysine linkages [12] | Very high | High (plate-based) [15] |
| Antibody-based | Variable | Dependent on antibody specificity [4] | High for specific targets | Medium |
A representative application involves using TUBEs to study RIPK2 (Receptor-Interacting Serine/Threonine-Protein Kinase 2) ubiquitination in inflammatory signaling and targeted degradation [13]. In this context:
- Inflammatory stimuli (L18-MDP) induce K63-linked ubiquitination of RIPK2, which can be captured specifically with K63-TUBEs or pan-TUBEs [13]
- RIPK2-directed PROTACs induce K48-linked ubiquitination, captured by K48-TUBEs and pan-TUBEs but not K63-TUBEs [13]
- TUBE-based assays can demonstrate inhibition of RIPK2 ubiquitination by small-molecule inhibitors like Ponatinib [13]
This approach provides a rapid, quantitative method for characterizing ubiquitin-mediated processes in drug development, facilitating the optimization of targeted protein degradation therapeutics.
Troubleshooting and Technical Considerations
Successful implementation of UBD-based ubiquitination studies requires attention to several technical considerations:
Sample Preparation Considerations:
- Always include deubiquitinase inhibitors (NEM or iodoacetamide) in lysis buffers to preserve ubiquitination signals [14] [13]
- Consider including proteasome inhibitors (MG132) when studying degradation targets [13]
- Use mild lysis conditions (avoid strong denaturants) for native interaction studies [14]
- Process samples quickly at 4°C to minimize deubiquitination and degradation [14]
Troubleshooting Common Issues:
- High Background: Increase wash stringency (salt concentration, detergent percentage) or optimize blocking conditions [15]
- Low Signal: Verify inhibitor activity, increase input protein, or try different UBD constructs with higher affinity [12]
- Inconsistent Results: Standardize lysis protocols and ensure consistent sample processing across experiments [14]
- Linkage Specificity Concerns: Include multiple controls with different chain-specific TUBEs and validate with known standards [13]
Experimental Design Recommendations:
- For discovery proteomics, use pan-specific UBDs (ThUBD, pan-TUBEs) to maximize coverage [12]
- For mechanistic studies, employ chain-specific TUBEs to determine linkage dependence [13]
- For drug discovery applications, implement plate-based ThUBD assays for high-throughput capability [15]
- Always include appropriate controls (untreated samples, specificity controls) for accurate interpretation [13]
The continuous development of engineered UBDs with enhanced affinity and reduced linkage bias, such as ThUBDs and optimized TUBEs, promises to further advance our ability to decipher the complex ubiquitin code and harness this knowledge for therapeutic applications [15] [12].
Ubiquitination is a crucial post-translational modification that regulates diverse cellular functions, including proteasomal degradation, signal transduction, DNA repair, and immune responses [13] [16]. This versatility stems from the complexity of ubiquitin conjugates, which can range from a single ubiquitin monomer (monoubiquitination) to polymers of various lengths and linkage types [16]. The ubiquitin-proteasome system (UPS) involves a cascade of E1, E2, and E3 enzymes that ultimately attach ubiquitin to substrate proteins, while deubiquitinases (DUBs) reverse this process [14] [16]. Among the eight distinct ubiquitin chain linkages, K48-linked chains primarily target proteins for proteasomal degradation, while K63-linked chains typically regulate non-proteolytic functions such as inflammatory signaling and protein trafficking [13] [17]. Traditional methods for studying ubiquitination, such as immunoblotting with ubiquitin antibodies or expression of epitope-tagged ubiquitin, present significant limitations including low throughput, potential artifacts, and an inability to preserve native cellular conditions [14] [6] [16]. The need to overcome these challenges prompted the development of Tandem Ubiquitin Binding Entities (TUBEs), engineered affinity reagents that have revolutionized the study of the ubiquitin code.
The TUBE Innovation: Conceptual and Technical Advancements
From Single UBA Domains to Tandem-Repeated Entities
The fundamental innovation of TUBEs lies in their engineered tandem arrangement of multiple Ubiquitin-Associated (UBA) domains within a single polypeptide chain [5] [6]. Individual UBA domains typically exhibit only micromolar affinity for ubiquitin, limiting their utility for efficient capture of ubiquitinated proteins from complex biological samples [16]. By combining multiple UBA domains in tandem, TUBEs achieve dramatically enhanced affinity for polyubiquitin chains, with dissociation constants (Kd) in the low nanomolar range (1-10 nM) [5] [6]. This architectural innovation transforms what would be weak individual interactions into a powerful, multivalent ubiquitin capture system capable of competing with endogenous ubiquitin receptors and protecting ubiquitinated proteins from degradation and deubiquitination [6] [18].
Key Advantages Over Traditional Methods
TUBEs offer several critical advantages that address the limitations of previous methodologies. Unlike epitope-tagged ubiquitin approaches, TUBEs require no genetic manipulation of cells or competition with endogenous ubiquitin, thereby preserving native ubiquitination landscapes [6] [18]. Compared to ubiquitin antibodies, TUBEs provide superior affinity and specificity while being more economical for large-scale studies [6]. Most notably, TUBEs protect polyubiquitin chains from disassembly by deubiquitinating enzymes (DUBs) and from proteasomal degradation, even in the absence of protease inhibitors normally required to block these activities [6] [18]. This protective function enables more accurate snapshotting of dynamic ubiquitination events under physiological conditions. Additionally, TUBEs can be engineered for pan-selective recognition of all ubiquitin chain types or for exquisite specificity toward particular linkages (e.g., K48, K63, or M1-linear chains), enabling researchers to decipher the functional consequences of specific ubiquitin signatures [5] [6].
TUBE Reagents and Experimental Implementation
Research Reagent Solutions
The successful implementation of TUBE-based methodologies relies on a suite of specialized reagents optimized for different experimental applications. The table below outlines key TUBE reagents and their specific research applications.
Table 1: Essential TUBE Reagents for Ubiquitin Research
| Reagent Type | Specific Examples | Key Features & Applications |
|---|---|---|
| Pan-Selective TUBEs | TUBE1, TUBE2 (e.g., UM401M, UM202) [13] [6] | Binds all polyubiquitin chain types with nanomolar affinity (1-10 nM); ideal for general ubiquitome enrichment and proteomic studies [6]. |
| Chain-Selective TUBEs | K48-TUBE, K63-TUBE, M1-TUBE [13] [6] [17] | Specifically recognizes K48-linked, K63-linked, or linear M1-linked polyubiquitin chains; enables linkage-specific functional studies [13] [17]. |
| TUBE-Conjugated Magnetic Beads | UM401M [13] [6] | TUBEs immobilized on magnetic beads; simplifies pulldown assays for enriching ubiquitinated proteins from cell lysates for Western blot or mass spectrometry [13] [6]. |
| Fluorophore-Labeled TUBEs | TAMRA-TUBE 2 (UM202) [6] | Contains a TAMRA fluorophore on the fusion tag for imaging applications; allows visualization of ubiquitin dynamics in cells without interfering with ubiquitin binding [6]. |
| TUBE-Coated Microplates | K48- and K63-TUBE coated plates [13] [17] | Enables development of high-throughput screening (HTS) assays in 96-well format for drug discovery applications, particularly for PROTAC characterization [13] [17]. |
Detailed Protocol: Investigating Linkage-Specific Ubiquitination of RIPK2 Using TUBEs
The following protocol details a specific application of TUBE technology to capture and analyze endogenous K63-linked ubiquitination of Receptor-Interacting Serine/Threonine-Protein Kinase 2 (RIPK2) in response to inflammatory stimulation in THP-1 cells [13] [17]. This methodology can be adapted for other target proteins and cellular contexts.
Background and Principle
Inflammatory signaling via the NOD2 pathway involves K63-linked ubiquitination of RIPK2 upon stimulation with muramyldipeptide (MDP) [13]. This protocol uses chain-specific TUBEs in a pulldown approach to selectively capture this modification, demonstrating how TUBEs can decipher context-dependent ubiquitination.
Materials and Reagents
- Cells: Human monocytic THP-1 cell line.
- Stimuli: L18-MDP (Lysine 18-muramyldipeptide, 200-500 ng/mL) to induce K63 ubiquitination of RIPK2 [13].
- Inhibitor: Ponatinib (100 nM), a RIPK2 inhibitor, for control experiments [13].
- Lysis Buffer: A specialized buffer optimized to preserve polyubiquitination, typically containing protease inhibitors and DUB inhibitors (e.g., N-ethylmaleimide (NEM)) [13].
- TUBE Reagents: Pan-selective TUBE-conjugated magnetic beads (e.g., UM401M) and/or chain-specific K63-TUBE beads [13] [6].
- Antibodies: Anti-RIPK2 antibody for immunoblotting detection.
Step-by-Step Procedure
Cell Culture and Treatment:
- Culture THP-1 cells under standard conditions.
- Pre-treat cells with either DMSO (vehicle control) or 100 nM Ponatinib for 30 minutes [13].
- Stimulate cells with either water (vehicle control) or 200-500 ng/mL L18-MDP for 30 minutes and 60 minutes to induce time-dependent K63 ubiquitination of RIPK2 [13].
Cell Lysis:
- Lyse cells using the pre-cooled, optimized lysis buffer. Maintain samples on ice to minimize deubiquitination and protein degradation.
- Clarify lysates by centrifugation at 14,000 × g for 15 minutes at 4°C.
- Quantify protein concentration in the supernatant using a compatible assay (e.g., BCA or Bradford assay).
TUBE Pulldown Enrichment:
- Incubate 50-100 µg of clarified cell lysate with Pan-selective or K63-TUBE conjugated magnetic beads for 2-4 hours at 4°C with gentle rotation [13].
- For high-throughput applications, as demonstrated in the recent study, lysates can be incubated in K63-TUBE or K48-TUBE coated 96-well plates [13] [17].
Washing and Elution:
- Wash beads extensively with ice-cold lysis buffer to remove non-specifically bound proteins.
- Elute bound ubiquitinated proteins by boiling in 1× SDS-PAGE loading buffer containing reducing agent (e.g., DTT) for 5-10 minutes.
Detection and Analysis:
- Resolve eluted proteins by SDS-PAGE and transfer to PVDF membrane.
- Probe the membrane with anti-RIPK2 antibody to detect ubiquitinated forms of RIPK2, which appear as higher molecular weight smears or discrete bands above the unmodified protein.
- As shown in the referenced study, the expected result is a strong signal for polyubiquitinated RIPK2 in L18-MDP stimulated samples captured by K63-TUBEs or Pan-TUBEs, but not by K48-TUBEs. Pre-treatment with Ponatinib should abrogate this signal [13].
The experimental workflow and the specific signaling pathway investigated in this protocol are illustrated below.
Figure 1: Experimental workflow for TUBE-based analysis of L18-MDP-induced K63 ubiquitination of RIPK2.
Application in Drug Discovery: Advancing PROTAC Characterization
TUBE technology has found a particularly impactful application in the field of targeted protein degradation (TPD), specifically in the characterization of Proteolysis Targeting Chimeras (PROTACs) and molecular glues [5] [13]. PROTACs are heterobifunctional small molecules that recruit E3 ubiquitin ligases to target proteins of interest, inducing their polyubiquitination and subsequent proteasomal degradation [13]. A critical step in evaluating PROTAC efficacy is confirming target protein ubiquitination, which has traditionally been challenging to assess for endogenous proteins in a high-throughput manner.
The integration of TUBEs into high-throughput screening (HTS) assays represents a significant advancement. As demonstrated in the recent study, chain-specific TUBEs can be coated onto microplates to create a capture surface for polyubiquitinated proteins from cell lysates [13] [17]. In this assay format, RIPK2 PROTAC (RIPK degrader-2) induced K48-linked ubiquitination of endogenous RIPK2, which was specifically captured by K48-TUBEs and Pan-TUBEs, but not by K63-TUBEs [13]. Conversely, L18-MDP-induced K63-linked ubiquitination was captured by K63-TUBEs and Pan-TUBEs, but not K48-TUBEs [13]. This ability to differentiate linkage-specific ubiquitination in a cellular context provides invaluable mechanistic insight during drug screening. The quantitative data from such TUBE-based HTS assays for RIPK2 are summarized below.
Table 2: Quantitative Performance of Chain-Selective TUBEs in Capturing Endogenous RIPK2 Ubiquitination [13] [17]
| Experimental Condition | TUBE Type Used for Capture | Relative RIPK2 Ubiquitination Signal | Biological Interpretation |
|---|---|---|---|
| L18-MDP Stimulation | K63-TUBE | Strong Signal | Inflammatory stimulus induces K63-linked ubiquitination of RIPK2 for signal transduction [13]. |
| L18-MDP Stimulation | K48-TUBE | Minimal/No Signal | Confirms specificity, showing L18-MDP does not induce K48-linked degradation signals [13]. |
| L18-MDP Stimulation | Pan-TUBE | Strong Signal | Pan-TUBE captures all ubiquitin linkages, validating overall ubiquitination [13]. |
| RIPK2 PROTAC Treatment | K48-TUBE | Strong Signal | PROTAC successfully induces K48-linked ubiquitination, targeting RIPK2 for degradation [13]. |
| RIPK2 PROTAC Treatment | K63-TUBE | Minimal/No Signal | Confirms PROTAC mechanism is specific to K48-linked degradation chains [13]. |
| RIPK2 PROTAC Treatment | Pan-TUBE | Strong Signal | Pan-TUBE confirms overall PROTAC-induced ubiquitination [13]. |
This TUBE-based platform overcomes the limitations of Western blotting (low throughput, semi-quantitative) and reporter assays (potential artifacts) [13]. It enables rapid, quantitative ranking of PROTAC potency based on their ability to induce ubiquitination of endogenous target proteins, thereby accelerating the drug discovery process for a wide range of diseases, including cancer, inflammatory disorders, and neurodegenerative conditions [5] [13]. The strategic use of different TUBE types in this process is outlined in the following workflow.
Figure 2: TUBE-based HTS assay application in PROTAC characterization. The assay uses K48- and Pan-TUBEs to confirm the specific ubiquitination induced by the heterobifunctional PROTAC molecule.
The innovation of Tandem Ubiquitin Binding Entities represents a paradigm shift in the study of ubiquitination. By transforming low-affinity single UBA domains into high-affinity, multivalent tools, TUBEs have overcome long-standing challenges in capturing, stabilizing, and characterizing the dynamic ubiquitinome. Their unique ability to protect polyubiquitin chains from deubiquitination and degradation under native conditions, coupled with the availability of chain-specific variants, has provided researchers with an unprecedented window into the functional complexity of the ubiquitin code. As evidenced by their critical role in advancing high-throughput screening for PROTAC discovery and other targeted therapies, TUBEs have firmly established themselves as indispensable reagents in both basic research and translational drug development, enabling scientists to decipher ubiquitin signaling with greater precision, efficiency, and physiological relevance than ever before.
Tandem Ubiquitin Binding Entities (TUBEs) represent a transformative technology in ubiquitin proteasome system (UPS) research, enabling unprecedented isolation and analysis of polyubiquitinated proteins. This application note details the molecular mechanisms underlying TUBEs' exceptional nanomolar affinity for polyubiquitin chains and their protective function against cellular degradation machinery. By harnessing multiple ubiquitin-binding domains (UBDs) in tandem, TUBEs achieve up to 1,000-fold higher affinity for polyubiquitin compared to single UBD domains while simultaneously shielding captured proteins from deubiquitinating enzymes (DUBs) and proteasomal degradation. Within drug discovery contexts, particularly in PROTAC development and targeted protein degradation, TUBEs provide crucial tools for validating compound efficacy and understanding ubiquitination dynamics. This document provides comprehensive protocols for implementing TUBE technology and quantitative data supporting its application in advanced proteomic studies.
The ubiquitin-proteasome system (UPS) represents a complex post-translational regulatory mechanism controlling nearly all cellular processes through targeted protein degradation and signaling. Research in this field has historically been challenging due to the labile nature of polyubiquitinated proteins, which are rapidly processed by deubiquitinating enzymes (DUBs) and the proteasome itself. Traditional methods relying on ubiquitin antibodies or overexpression of tagged ubiquitin often yield insufficient affinity and specificity while potentially introducing experimental artifacts [6].
Tandem Ubiquitin Binding Entities (TUBEs) were engineered to overcome these limitations through a sophisticated protein design approach. By linking multiple ubiquitin-binding domains (UBDs) in sequence, TUBEs create an avidity effect that dramatically enhances binding capability toward polyubiquitin chains [6] [5]. This architectural innovation enables researchers to capture, stabilize, and characterize polyubiquitinated proteins at physiological levels without requiring proteasome inhibitors or genetic manipulation of cellular systems.
The significance of TUBE technology extends beyond basic research into drug discovery, particularly in the rapidly expanding field of targeted protein degradation. As PROTACs (Proteolysis-Targeting Chimeras) and molecular glues emerge as promising therapeutic modalities, TUBEs provide essential tools for validating target engagement and understanding the mechanism of action of these novel compounds [5] [19]. The ability to precisely monitor ubiquitination events in live cells using TUBE-based assays represents a critical advancement for high-throughput screening in pharmaceutical development.
Molecular Mechanisms of Action
Structural Basis for High-Affinity Binding
The exceptional binding capability of TUBEs originates from their strategic assembly of multiple ubiquitin-binding domains (UBDs) in a single polypeptide chain. Each UBD maintains the intrinsic ubiquitin recognition function, but when positioned in tandem, they create a cooperative binding effect that dramatically enhances overall affinity for polyubiquitin chains. This architectural innovation results in equilibrium dissociation constants (Kd) in the nanomolar range (typically 1-10 nM), representing a 100 to 1,000-fold increase in affinity compared to isolated UBD domains [6] [5].
The molecular mechanism involves simultaneous engagement with multiple ubiquitin subunits within a polyubiquitin chain. Whereas single UBDs interact with limited contact points, tandemly arranged UBDs span a broader surface area of the polyubiquitin chain, forming multiple non-covalent interactions that collectively produce an avidity effect far exceeding monomeric binding capability [20]. This multi-point attachment creates a remarkably stable complex that resists dissociation during experimental procedures, enabling reliable capture of polyubiquitinated proteins even at low abundance.
Protection Against Cellular Degradation Machinery
Beyond their exceptional binding properties, TUBEs provide crucial protective functions that stabilize polyubiquitinated proteins against the cellular machinery that would normally process them. The binding mechanism physically shields polyubiquitin chains from the catalytic sites of deubiquitinating enzymes (DUBs), thereby preventing chain editing or complete deubiquitination [6]. This protective effect maintains the ubiquitination status of captured proteins throughout isolation procedures, preserving their native state for downstream analysis.
Additionally, TUBEs sterically hinder recognition by proteasomal subunits that typically initiate degradation of polyubiquitinated substrates. This protection occurs even in the absence of proteasome inhibitors traditionally required to stabilize ubiquitinated proteins [6] [20]. The concurrent inhibition of both deubiquitination and proteasomal degradation allows researchers to detect and characterize polyubiquitinated proteins that would otherwise be too transient for comprehensive analysis, substantially expanding the experimental accessibility of the ubiquitinome.
Linkage-Specific Recognition
Advanced TUBE designs incorporate UBD variants with selective preferences for specific ubiquitin chain linkages, enabling researchers to target particular ubiquitin signaling pathways. For example, M1-linked linear ubiquitin chains can be specifically isolated using specialized TUBEs with 1,000 to 10,000-fold preference for M1 chains over K48 or K63 linkages [21]. Similarly, K48- and K63-specific TUBEs permit selective investigation of degradation signals versus non-degradative ubiquitin signaling.
This linkage selectivity arises from precise molecular complementarity between the UBD arrangements and the distinct three-dimensional architectures adopted by different polyubiquitin chain types. The structural basis for this discrimination enables researchers to dissect the complex biological functions associated with specific ubiquitin linkages, moving beyond bulk polyubiquitin analysis to pathway-specific investigation [21] [5].
Table 1: Affinity Characteristics of Select TUBE Reagents
| TUBE Type | Specificity | Dissociation Constant (Kd) | Selectivity Ratio | Applications |
|---|---|---|---|---|
| M1 TUBE | Linear (M1) chains | Nanomolar range | 1,000-10,000x preference over K48/K63 | NF-κB signaling, inflammation research |
| K48 TUBE | K48-linked chains | Nanomolar range | Selective for degradation signals | Proteasomal degradation studies |
| K63 TUBE | K63-linked chains | Nanomolar range | Selective for non-degradative signals | DNA repair, signaling complexes |
| TUBE 2 | Pan-selective | 1-10 nM | Equivalent affinity for K48/K63 | General ubiquitome analysis |
Quantitative Affinity and Selectivity Data
The performance advantages of TUBEs are substantiated by rigorous quantitative measurements of their binding characteristics. Pan-selective TUBEs, such as TUBE 2, exhibit consistent low nanomolar affinity (Kd = 1-10 nM) for diverse polyubiquitin chain types, enabling comprehensive ubiquitome profiling without linkage bias [6]. This uniform high affinity ensures efficient capture of polyubiquitinated proteins regardless of chain topology, making pan-selective TUBEs ideal for initial discovery-phase experiments.
For investigations targeting specific ubiquitin-dependent pathways, linkage-specific TUBEs provide exceptional discriminatory capability. The M1-linear TUBE demonstrates a remarkable 10,000-fold preference for M1-linked chains over K48- or K63-linked alternatives, achieving selective isolation of proteins modified by linear ubiquitination [21]. This extraordinary selectivity enables precise interrogation of inflammatory signaling pathways regulated by linear ubiquitin assembly complex (LUBAC)-mediated ubiquitination.
The quantitative binding superiority of TUBEs becomes particularly evident when compared to traditional ubiquitin-binding reagents. While conventional ubiquitin antibodies often exhibit micromolar affinities and significant cross-reactivity with unrelated antigens, TUBEs provide 100 to 1,000-fold higher affinity while maintaining exceptional specificity for polyubiquitin chains over monoubiquitin [20]. This performance advantage translates directly to enhanced sensitivity in detecting low-abundance polyubiquitinated species and reduced background in pull-down experiments.
Table 2: Protective Functions of TUBE Technology
| Protective Function | Mechanism | Experimental Benefit |
|---|---|---|
| Deubiquitination protection | Steric hindrance of DUB catalytic sites | Preserves ubiquitination state during processing |
| Proteasomal shielding | Physical blocking of proteasome recognition | Eliminates need for proteasome inhibitors |
| General stabilization | Structural support to ubiquitin-protein linkage | Enables study of labile ubiquitination events |
| Background reduction | High affinity minimizes non-specific binding | Enhances signal-to-noise ratio in detection |
Research Applications and Experimental Implementation
Ubiquitinated Protein Enrichment and Proteomics
The primary application of TUBE technology involves efficient isolation of polyubiquitinated proteins for proteomic analysis and identification of novel ubiquitination substrates. The exceptional affinity and protective functions of TUBEs make them particularly valuable for capturing transient or low-abundance ubiquitinated species that evade detection with conventional methods. When conjugated to solid supports such as magnetic beads, TUBEs enable single-step purification of polyubiquitinated complexes without centrifugation, facilitating complete supernatant removal and significantly reducing background [22].
This application proves especially powerful in biomarker discovery and disease mechanism studies, where comprehensive ubiquitinome profiling can reveal pathway-specific alterations in ubiquitination patterns. The compatibility of TUBE-based isolations with multiple detection platforms—including Western blotting, mass spectrometry, and luminescence-based assays—provides researchers with exceptional methodological flexibility [5] [19]. For drug discovery programs, this approach enables quantitative assessment of compound-induced changes in global ubiquitination patterns, facilitating mechanism of action studies for UPS-targeting therapeutics.
High-Throughput Screening and Drug Discovery
TUBE technology has been successfully adapted to high-throughput screening formats that accelerate the identification and characterization of UPS-modulating compounds. By combining TUBEs with luminescence reporting systems such as NanoBiT, researchers have developed live-cell assays capable of monitoring substrate ubiquitination kinetics in real time [19]. These platforms enable rapid quantification of GSPT1 ubiquitination in response to PROTAC treatment, establishing rank-order potency for compound libraries and supporting structure-activity relationship studies.
In the context of molecular glue development, TUBE-based assays provide critical functional data on target protein ubiquitination that complements traditional viability and degradation readouts. The ability to directly measure the ubiquitination events preceding protein degradation offers valuable insights into compound mechanism and efficiency, informing medicinal chemistry optimization efforts [5]. Furthermore, the compatibility of these assays with automation makes them ideally suited for the high-throughput screening campaigns necessary to advance targeted protein degradation therapeutics.
Validation of PROTAC Efficiency
PROTACs (Proteolysis-Targeting Chimeras) represent a promising therapeutic modality that redirects E3 ubiquitin ligase activity to neo-substrates, inducing their polyubiquitination and subsequent degradation. TUBEs provide essential tools for confirming successful target engagement by directly demonstrating polyubiquitination of the protein of interest following PROTAC treatment [5]. This application is particularly valuable for establishing proof-of-concept for novel PROTAC designs and optimizing linker chemistry to maximize ubiquitination efficiency.
The chain-type selectivity of certain TUBE variants enables researchers to determine the specific ubiquitin linkage topology generated by PROTAC-induced ubiquitination, offering insights into the mechanism of E3 ligase recruitment and the efficiency of subsequent degradation. This capability to discriminate between productive (typically K48-linked) and non-productive ubiquitination events helps guide PROTAC optimization toward compounds that generate degradation-competent ubiquitin signatures [5].
Detailed Experimental Protocols
Magnetic Bead-Based Pulldown of Polyubiquitinated Proteins
Principle: This protocol utilizes TUBE 2 conjugated to magnetic beads for efficient isolation of polyubiquitinated proteins from cell and tissue extracts without centrifugation, enabling complete supernatant removal for reduced background [22].
Reagents and Solutions:
- Lysis Buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 1 mM EDTA, supplemented with protease inhibitors (optional)
- TUBE 2 Magnetic Beads (LifeSensors UM402M)
- Wash Buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% NP-40
- Elution Buffer: 0.2 M glycine-HCl (pH 2.5) or 1X SDS-PAGE loading buffer
Procedure:
- Cell Lysis: Pre-chill lysis buffer to 4°C. Wash cells appropriately and add 500 μL lysis buffer per 10 cm culture dish (approximately 1.5×10⁶ cells). Collect cells by scraping and transfer to pre-chilled microcentrifuge tube.
- Lysate Clarification: Incubate lysate on ice for 15 minutes, then clarify by centrifugation at 14,000×g for 10 minutes at 4°C. Transfer supernatant to a new tube.
- TUBE Incubation: Add 10-20 μL of equilibrated TUBE 2 magnetic beads to the lysate. Incubate for 4 hours at 4°C with gentle agitation.
- Bead Capture: Place tube on magnetic separator for 1-2 minutes until solution clears. Carefully remove and discard supernatant.
- Washing: Resuspend beads in 500 μL wash buffer, incubate for 5 minutes at 4°C with agitation, and capture on magnetic separator. Discard supernatant. Repeat wash step twice.
- Elution: Add 30-50 μL elution buffer to beads and incubate for 1 hour at 4°C with agitation. Capture beads on magnetic separator and transfer supernatant containing eluted polyubiquitinated proteins to a new tube.
- Neutralization: For acidic elutions, neutralize with 1/10 volume 1 M Tris-HCl (pH 8.0). Samples can be analyzed immediately by Western blot or mass spectrometry.
Technical Notes:
- For optimal results, maintain consistent temperature (4°C) throughout the procedure
- Protein yield can be increased by extending incubation time with TUBE beads to overnight
- Avoid freeze-thaw cycles with TUBE reagents; store according to manufacturer specifications
- For proteomic applications, consider on-bead digestion to minimize contamination
Live-Cell Ubiquitination Monitoring Assay
Principle: This protocol adapts TUBE technology for real-time monitoring of substrate ubiquitination in live cells by combining TUBEs with NanoBiT luminescence technology, enabling high-throughput compound screening [19].
Reagents and Solutions:
- NanoBiT Ubiquitination Substrate Construct
- TUBE Fusion Protein appropriate for detection system
- Compound library for screening
- Cell culture medium appropriate for cell line
- Luminescence detection reagents
Procedure:
- Cell Preparation: Seed cells in appropriate multi-well plates for luminescence reading and incubate overnight to reach 70-80% confluence.
- Transfection: Co-transfect cells with NanoBiT-tagged substrate of interest and TUBE fusion construct using preferred transfection method. Incubate for 24-48 hours to allow expression.
- Compound Treatment: Add PROTACs, molecular glues, or other test compounds at desired concentrations. Include appropriate controls (DMSO vehicle, known activators/inhibitors).
- Signal Detection: Measure luminescence at predetermined timepoints following compound addition using plate reader capable of luminescence detection.
- Data Analysis: Normalize luminescence values to vehicle controls and calculate fold-change in ubiquitination relative to baseline.
Technical Notes:
- Optimize transfection efficiency for consistent results across experimental replicates
- Determine optimal reading timepoints through kinetic studies before large-scale screening
- Include controls for non-specific luminescence and auto-ubiquitination background
- Adapt protocol for specific substrates by modifying TUBE specificity and detection parameters
Research Reagent Solutions
The effective implementation of TUBE technology requires appropriate selection from available reagent formats, each offering distinct advantages for specific applications. The table below summarizes key TUBE products and their recommended research uses.
Table 3: Essential TUBE Reagents for Ubiquitin Research
| Product Name | Tag/Conjugate | Specificity | Key Features | Applications |
|---|---|---|---|---|
| UM206: M1 Linear TUBE | His6 | Linear (M1) chains | 1,000-10,000x preference for M1 chains | NF-κB signaling, inflammation studies |
| UM402M: TUBE 2 Magnetic Beads | Magnetic beads | Pan-selective | No centrifugation needed, low background | High-throughput pulldowns, proteomics |
| UM604: K63 TUBE | FLAG | K63-linked chains | Selective for non-degradative ubiquitination | DNA repair, kinase signaling studies |
| UM507T: K48 TUBE | TAMRA | K48-linked chains | Fluorescent detection of degradation signals | PROTAC validation, degradation imaging |
| UM201: TUBE 1 | His6 | Moderate K63 preference | 10x higher affinity for K63 vs K48 chains | Pathway-specific ubiquitination studies |
| UM301: Biotin-TUBE 1 | Biotin | Moderate K63 preference | Compatible with streptavidin systems | Far-Western blotting, detection |
Schematic Representation of TUBE Mechanisms
The following diagram illustrates the molecular mechanism by which TUBEs achieve high-affinity binding to polyubiquitin chains while providing protection from deubiquitination and proteasomal recognition:
Schematic of TUBE Molecular Mechanism
This diagram illustrates how the tandem arrangement of ubiquitin-binding domains (UBDs) in TUBEs enables simultaneous engagement with multiple ubiquitin subunits within a polyubiquitin chain, creating an avidity effect that produces nanomolar affinity. The comprehensive binding coverage physically blocks access by deubiquitinating enzymes (DUBs) and proteasomal recognition elements, thereby stabilizing polyubiquitinated proteins against degradation and editing.
TUBE technology represents a paradigm shift in ubiquitin research, providing unprecedented capability to capture, stabilize, and analyze polyubiquitinated proteins through elegantly engineered molecular mechanisms. The tandem arrangement of ubiquitin-binding domains generates exceptional binding affinity in the nanomolar range while simultaneously protecting substrates from cellular degradation machinery. These properties make TUBEs indispensable tools for both basic research investigating ubiquitin-dependent processes and drug discovery programs developing targeted protein degradation therapeutics. As the ubiquitin field continues to expand, TUBE-based methodologies offer robust, reproducible platforms for deciphering the complex biological functions of ubiquitination in health and disease.
Tandem Ubiquitin Binding Entities (TUBEs) are engineered, high-affinity reagents composed of multiple ubiquitin-associated (UBA) domains that bind polyubiquitin chains with significantly higher affinity than monomeric UBAs [23]. These tools have emerged as indispensable in ubiquitin research, enabling the isolation, enrichment, and characterization of polyubiquitinated proteins from native systems such as cell lines and tissues under conditions that protect ubiquitin chains from deubiquitinases (DUBs) and the proteasome, even in the absence of standard inhibitors [24]. The functional consequences of protein polyubiquitination are primarily determined by the type of ubiquitin chain assembled on the substrate [23]. Among the eight distinct types of ubiquitin chains, lysine 48 (K48)-linked chains are specifically associated with proteasomal degradation, while lysine 63 (K63)-linked chains are primarily involved in regulating signal transduction, protein trafficking, and immune responses [25] [13]. Linear (M1-linked) chains also play crucial roles in inflammatory and immune signaling pathways. This linkage specificity forms the basis of the "ubiquitin code," which TUBEs are designed to decipher [26].
Pan-selective TUBEs bind to all polyubiquitin chain linkage types without discrimination, making them appropriate when the linkage type is unknown or when a broad enrichment of ubiquitinated proteins is desired [24]. In contrast, linkage-selective TUBEs, such as those specific for K48, K63, or M1 linkages, are engineered for enhanced selectivity toward particular chain architectures, enabling researchers to investigate the specific polyubiquitin chain linkage on their substrate protein [23]. This specificity is crucial because different ubiquitin linkages dictate distinct functional outcomes in cellular processes. The development of linkage-specific TUBEs represents a significant advancement over traditional methods like ubiquitin antibodies, offering nanomolar affinities and up to 100-fold preference for their target polyubiquitin chains over other linkages [23] [24].
Quantitative Data on TUBE Binding Properties
The utility of TUBEs in experimental applications is rooted in their quantitative binding characteristics. The table below summarizes the affinity and specificity data for major TUBE types:
Table 1: Affinity and Specificity Profiles of Select TUBEs
| TUBE Type | Target Linkage | Affinity (Kd) | Specificity Over Other Linkages | Primary Applications |
|---|---|---|---|---|
| K48 TUBE HF | K48-linked polyUb | ~20 nM [23] [24] | >100-fold [24] (>2 µM for other linkages [23]) | Enrichment of proteins targeted for proteasomal degradation [23] |
| K63 TUBE | K63-linked polyUb | Single-digit nM range [24] | Information missing | Studying inflammatory signaling, protein trafficking, and DNA repair pathways [25] |
| M1 TUBE | Linear polyUb | Information missing | Information missing | Research on NF-κB and immune signaling pathways |
| Pan-TUBE (TUBE1/2) | All linkages | Single-digit nM range [24] | No linkage discrimination [24] | General ubiquitome enrichment when linkage is unknown; DUB protection [24] |
The high fidelity of these tools enables precise experimental outcomes. For instance, K48 TUBE HF binds with nanomolar affinity for K48 polyubiquitin chains, demonstrating higher affinity than most commercially available ubiquitin antibodies [23]. This performance allows for quantitative enrichment of ubiquitylated proteins from tissues and cells without requiring SILAC labeling for mass spectrometry proteomics applications [23].
The application of chain-specific TUBEs in high-throughput screening (HTS) assays enables investigation of ubiquitination dynamics on endogenous proteins. Recent research has demonstrated that chain-selective TUBEs can differentiate context-dependent linkage-specific ubiquitination, as shown in the study of RIPK2, where K63-TUBEs captured inflammation-associated ubiquitination while K48-TUBEs identified PROTAC-induced degradation signaling [25] [13]. This specificity is quantifiable, with K63-TUBEs and Pan-TUBEs capturing L18-MDP-stimulated K63 ubiquitination of RIPK2, while K48-TUBEs showed no appreciable signal for this pathway [25].
Experimental Protocols for Linkage-Specific Ubiquitination Assessment
Protocol 1: Enrichment of Linkage-Specific Polyubiquitinated Proteins
This protocol describes the use of linkage-specific TUBEs for isolating and enriching polyubiquitinated proteins with specific chain linkages from cell extracts. The procedure typically utilizes TUBEs tagged with His6 or biotin and immobilized on appropriate affinity resins [23] [24].
Materials:
- TUBE Reagents: Linkage-specific TUBEs (e.g., K48 TUBE HF [His6], K63-TUBE, M1-TUBE) [23] [24]
- Cell Lysate: Prepared using lysis buffer optimized to preserve polyubiquitination (e.g., containing DUB inhibitors) [25]
- Binding Buffer: Compatible with TUBE-ubiquitin interaction
- Elution Buffer: Typically containing SDS or competitive elution agents
- Affinity Resin: Nickel-NTA resin for His6-tagged TUBEs or streptavidin resin for biotinylated TUBEs [24]
Procedure:
- Prepare Cell Lysate: Lyse cells in an appropriate buffer supplemented with DUB inhibitors to prevent chain disassembly. Maintain samples on ice throughout the process. The lysis buffer should be optimized to preserve polyubiquitination states [25].
- Immobilize TUBEs: For His6-tagged TUBEs, incubate with nickel-NTA resin. For biotinylated TUBEs, incubate with streptavidin resin. Use control uncoupled agarose beads as a negative control [24].
- Incubate Lysate with TUBE-Resin Complex: Mix cell lysate (typically 500 µg to 1 mg total protein) with TUBE-bound resin. Incubate for 2-4 hours at 4°C with gentle rotation.
- Wash Beads: Perform multiple washes with binding buffer to remove non-specifically bound proteins.
- Elute Bound Proteins: Elute polyubiquitinated proteins using SDS-PAGE sample buffer or competitive elution with free ubiquitin. Analyze by Western blotting or mass spectrometry.
Applications: This protocol is ideal for isolating K48-polyubiquitinated proteins for proteomic studies, Far-Western detection of specific polyubiquitinated proteins, and enrichment of linkage-specific ubiquitylated proteins from cell and tissue extracts [23] [24].
Protocol 2: Assessing Linkage-Specific Ubiquitination of Endogenous Proteins in High-Throughput Format
This protocol adapts TUBE technology for high-throughput screening (HTS) applications, enabling the assessment of PROTAC or molecular glue-mediated endogenous target protein ubiquitination in a linkage-specific manner [25].
Materials:
- Chain-Specific TUBEs: K48-TUBEs, K63-TUBEs, and Pan-TUBEs
- 96-Well Plates: Coated with appropriate TUBE capture reagents
- Cell Culture: Appropriate cell line for studied pathway (e.g., THP-1 cells for inflammatory signaling)
- Stimuli/Inhibitors: Context-specific agents (e.g., L18-MDP for K63 ubiquitination, PROTACs for K48 ubiquitination, Ponatinib for RIPK2 inhibition) [25]
- Detection Antibodies: Target protein-specific antibodies
Procedure:
- Cell Treatment: Treat cells with appropriate stimuli to induce specific ubiquitination events. For example:
- Cell Lysis: Lyse cells using optimized lysis buffer that preserves polyubiquitination.
- Capture with TUBEs: Transfer lysates to 96-well plates coated with linkage-specific TUBEs (K48-TUBEs, K63-TUBEs, or Pan-TUBEs). Incubate to allow binding.
- Wash and Detect: Wash plates and detect captured ubiquitinated proteins using target-specific antibodies.
Applications: This HTS-compatible protocol enables quantitative analysis of ubiquitin linkage diversity in response to various stimuli and assessment of PROTAC-mediated target ubiquitination, providing a more physiologically relevant screening platform compared to traditional methods like Western blotting [25].
Signaling Pathways and Experimental Workflows
The relationship between ubiquitin chain types and their cellular functions, along with experimental workflows for their study, can be visualized through the following pathways and protocols:
Cellular Functions of Ubiquitin Linkages and Modulators
The experimental workflow for TUBE-based analysis of linkage-specific ubiquitination involves multiple stages from sample preparation to detection:
TUBE-Based Ubiquitin Enrichment Workflow
The Scientist's Toolkit: Essential Research Reagents
Table 2: Essential Research Reagents for TUBE-Based Ubiquitination Studies
| Reagent/Tool | Specific Example | Function | Key Features |
|---|---|---|---|
| K48 TUBE HF | LifeSensors UM207 (His6-tagged) [24] | Selective isolation/enrichment of K48-linked polyubiquitinated proteins | ~20 nM affinity for K48 chains; >100-fold specificity over other linkages [23] [24] |
| K63 TUBE | LifeSensors K63 TUBE | Selective isolation/enrichment of K63-linked polyubiquitinated proteins | Single-digit nM affinity for K63 chains [24] |
| M1 TUBE | LifeSensors UM306 (Biotinylated) [24] | Selective isolation/enrichment of linear (M1-linked) polyubiquitin chains | High affinity/selectivity for M1 linkages |
| Pan-TUBE | LifeSensors TUBE1/TUBE2 [24] | General enrichment of polyubiquitinated proteins regardless of linkage | Single-digit nM affinity for all chain types; strong DUB protection [24] |
| Control Beads | LifeSensors UM400 (uncoupled agarose beads) [24] | Negative control for ubiquitin-related assays | Ensures specificity in pull-down experiments |
| DUB Inhibitors | N-ethylmaleimide (NEM), Chloroacetamide (CAA) [26] | Prevent ubiquitin chain disassembly during processing | Cysteine alkylators that target DUBs; choice affects Ub-binding interactions [26] |
This toolkit enables researchers to design experiments with appropriate controls and specificities for deciphering the ubiquitin code in various biological contexts. The selection of specific TUBEs should be guided by the research question—whether investigating general ubiquitination (Pan-TUBEs) or specific functional outcomes associated with particular linkages (chain-specific TUBEs).
A Practical Protocol: From Cell Lysates to Ubiquitinated Protein Enrichment with TUBEs
Protein ubiquitination is a critical post-translational modification that regulates virtually all aspects of eukaryotic cell biology, influencing diverse cellular functions including proteasomal degradation, signal transduction, DNA repair, and immune responses [13] [27]. The remarkable functional diversity of ubiquitination stems from the structural complexity of ubiquitin itself—a 76-amino acid protein that can form polymer chains through eight distinct linkage types (M1, K6, K11, K27, K29, K33, K48, and K63) via its internal lysine residues and N-terminal methionine [27] [16]. The specific linkage type determines the three-dimensional architecture of the polyubiquitin chain and ultimately dictates the cellular outcome for the modified substrate, creating what is known as the "Ubiquitin Code" [27].
Tandem Ubiquitin Binding Entities (TUBEs) represent a breakthrough technology for deciphering this complex ubiquitin code. These engineered reagents consist of multiple ubiquitin-binding domains (UBDs) connected in tandem, conferring nanomolar affinities for polyubiquitin chains—significantly higher than single UBDs [13] [16]. This enhanced affinity enables TUBEs to protect ubiquitin chains from deubiquitinating enzymes (DUBs) during cell lysis and experimental procedures, preserving the native ubiquitination status of proteins [28]. TUBEs are broadly categorized into two classes: pan-specific TUBEs that recognize all ubiquitin linkage types with high affinity, and linkage-specific TUBEs that selectively bind to particular chain architectures such as K48 or K63 linkages [13] [23]. The strategic selection between these TUBE classes is paramount for experimental success, as each offers distinct advantages tailored to different research objectives in ubiquitin proteomics and drug discovery.
Understanding Polyubiquitin Chain Linkages and Functions
The functional consequences of protein polyubiquitination are primarily determined by the specific linkage type of the ubiquitin chain assembled on the substrate protein [27] [23]. Among the eight canonical linkage types, K48-linked and K63-linked chains are the most abundant and best characterized, collectively constituting approximately 70% of cellular ubiquitin linkages [27]. K48-linked polyubiquitin chains are the principal signal for targeting substrate proteins to the 26S proteasome for degradation, serving as a central mechanism in maintaining cellular proteostasis [13] [16]. In contrast, K63-linked polyubiquitin chains primarily function in non-proteolytic signaling pathways, regulating processes such as inflammatory signaling through NF-κB activation, DNA damage repair, protein trafficking, and kinase activation [13] [27].
The remaining "atypical" linkage types (M1, K6, K11, K27, K29, and K33) are less abundant and their functions are still being elucidated, though they are known to play important roles in cell cycle regulation, proteotoxic stress responses, and immune signaling [27]. The different polyubiquitin linkage types adopt distinct structures and conformations, creating unique interaction surfaces that are recognized by specific ubiquitin-binding proteins to mediate diverse cellular outcomes [27]. This linkage-specific functionality underscores the critical importance of experimental tools that can distinguish between different ubiquitin chain types when investigating ubiquitin-dependent processes.
Table 1: Major Ubiquitin Linkage Types and Their Primary Cellular Functions
| Linkage Type | Relative Abundance | Primary Cellular Functions | Key Characteristics |
|---|---|---|---|
| K48-linked | ~40% (Most abundant) | Proteasomal degradation [13] [16] | Canonical degradation signal |
| K63-linked | ~30% (Second most abundant) | Non-proteolytic signaling, NF-κB activation, DNA repair, protein trafficking [13] [27] | Forms extended chains with distinct interfaces |
| M1-linked (Linear) | Low | NF-κB activation, immune signaling [27] | Generated by LUBAC complex |
| K11-linked | Low | ER-associated degradation, cell cycle regulation [27] | Similar structure to K48-linked chains |
| K29/K33-linked | Low | Less characterized roles in signaling [27] | Atypical linkage types |
Pan-Specific vs. Linkage-Specific TUBEs: A Comparative Analysis
The decision between pan-specific and linkage-specific TUBEs represents a fundamental strategic choice in experimental design, with each approach offering distinct advantages and limitations. Pan-specific TUBEs are engineered to recognize a common structural feature present in all ubiquitin linkage types, typically binding with high affinity (nanomolar range) to diverse polyubiquitin chains regardless of their linkage specificity [13] [16]. These reagents are particularly valuable for global ubiquitome analysis, discovery-based proteomics, and applications requiring comprehensive capture of the total ubiquitinated proteome. Their broad specificity makes them ideal for initial characterization of ubiquitination events when the linkage type is unknown, or when studying proteins that may be modified with multiple chain types under different cellular conditions [13].
In contrast, linkage-specific TUBEs are designed with exquisite selectivity for particular ubiquitin chain architectures. For example, the K48-TUBE HF (High Fidelity) exhibits approximately 100-fold greater selectivity for K48-linked chains (~20 nM affinity) over other linkage types (>2 µM affinity) [23]. Similarly, K63-specific and M1-linear-specific TUBEs have been developed to selectively enrich their respective chain types [13] [23]. These reagents are indispensable for investigating the functional consequences of specific ubiquitination events, such as distinguishing between proteasomal targeting (K48-linked) and signaling functions (K63-linked) [13]. They enable researchers to decipher the physiological context of ubiquitin signaling, as demonstrated in studies of RIPK2, where K63-linked ubiquitination activates inflammatory signaling while K48-linked ubiquitination targets the protein for degradation [13].
Table 2: Comparison of Pan-Specific and Linkage-Specific TUBE Applications
| Parameter | Pan-Specific TUBEs | Linkage-Specific TUBEs |
|---|---|---|
| Binding Scope | Broad recognition of all ubiquitin linkage types [16] | Selective for specific linkages (e.g., K48, K63, M1) [13] [23] |
| Primary Applications | Global ubiquitome analysis, proteomic discovery, initial characterization [28] [16] | Functional studies of specific pathways, distinguishing degradation vs. signaling events [13] |
| Key Advantages | Comprehensive enrichment, unknown linkage scenarios, high sensitivity [16] | Functional insights, pathway-specific analysis, reduced background [13] [23] |
| Limitations | Cannot distinguish linkage types, may capture irrelevant ubiquitinated proteins [13] | May miss alternative linkages, requires prior knowledge or hypothesis [13] |
| Optimal Use Cases | Proteomic screens, identifying novel ubiquitination events, total ubiquitin quantification [28] | Investigating specific biological processes (e.g., inflammation, degradation), validating linkage-dependent functions [13] |
Decision Framework: Selecting the Appropriate TUBE for Your Research
The selection between pan-specific and linkage-specific TUBEs should be guided by the primary research objective, existing knowledge about the target protein, and the specific biological question under investigation. The following decision framework provides a systematic approach to TUBE selection:
When to Use Pan-Specific TUBEs
Discovery-phase research represents the primary application for pan-specific TUBEs. When investigating previously uncharacterized proteins or conducting unbiased ubiquitome profiling, pan-specific TUBEs offer the broadest capture capability, ensuring that all potential ubiquitination events are enriched regardless of linkage type [28] [16]. This approach was successfully employed in an integrative analysis of the ubiquitin proteome from MCF7 breast cancer cells, where pan-specific TUBEs identified 643 ubiquitinated proteins involved in diverse cellular processes including protein synthesis, cellular transport, and stress response pathways [28].
Total ubiquitination assessment represents another key application, particularly when quantifying global changes in ubiquitination in response to cellular stimuli, pharmacological treatments, or pathological conditions. For example, when evaluating the effect of proteasome inhibitors or E3 ligase modulators, pan-specific TUBEs provide a comprehensive view of alterations across the entire ubiquitinated proteome [16]. Additionally, protein interaction studies aimed at identifying complexes associated with ubiquitinated proteins benefit from pan-specific TUBEs, as they preserve the native ubiquitin landscape and facilitate the co-purification of ubiquitin-binding proteins and associated complexes [28].
When to Use Linkage-Specific TUBEs
Functional mechanism studies represent the strongest rationale for selecting linkage-specific TUBEs. When investigating whether ubiquitination serves degradative versus non-degradative functions, the simultaneous use of K48-specific and K63-specific TUBEs can provide definitive discrimination [13]. This approach was elegantly demonstrated in research on RIPK2, where inflammatory stimulus (L18-MDP) induced K63-linked ubiquitination captured specifically by K63-TUBEs, while a PROTAC degrader induced K48-linked ubiquitination selectively enriched by K48-TUBEs [13].
Pathway-specific investigations focused on biological processes known to involve specific ubiquitin linkages represent another key application. For example, studies of NF-κB signaling typically involve both K63-linked and M1-linear ubiquitination events, making linkage-specific TUBEs essential tools for dissecting the contribution of each chain type to pathway activation [13] [27]. Similarly, therapeutic development applications, particularly in the growing field of targeted protein degradation using PROTACs and molecular glues, benefit immensely from K48-specific TUBEs that can specifically monitor the formation of degradative ubiquitin chains on target proteins [13].
Integrated Experimental Approaches
For comprehensive ubiquitination characterization, a sequential or parallel approach utilizing both pan-specific and linkage-specific TUBEs often provides the most complete insight. Initial broad profiling with pan-specific TUBEs can identify ubiquitination events of interest, followed by linkage-specific analysis to determine the functional consequences. This integrated strategy leverages the strengths of both reagent classes while mitigating their individual limitations.
Detailed Methodologies for TUBE-Based Ubiquitination Analysis
Protocol for Linkage-Specific Ubiquitination Analysis Using TUBEs
The following detailed protocol describes the specific application of chain-selective TUBEs to differentiate context-dependent ubiquitination of endogenous proteins, based on methodologies successfully used to investigate RIPK2 ubiquitination dynamics [13]:
Cell Lysis and Preparation:
- Culture cells under experimental conditions (e.g., THP-1 human monocytic cells for inflammatory signaling studies).
- Prepare lysis buffer optimized to preserve polyubiquitination: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 1 mM EDTA, supplemented with fresh protease inhibitors (including 10 mM N-ethylmaleimide to inhibit DUBs) and phosphatase inhibitors [13].
- Treat cells according to experimental design. For RIPK2 studies: pre-treat with either DMSO vehicle or inhibitor (e.g., 100 nM Ponatinib) for 30 minutes, followed by stimulation with 200-500 ng/ml L18-MDP (for K63 ubiquitination) or PROTAC (for K48 ubiquitination) for 30-60 minutes [13].
- Lyse cells using 1 mL buffer per 10-20 million cells, incubate on ice for 30 minutes with occasional vortexing, then centrifuge at 16,000 × g for 15 minutes at 4°C to clear lysates.
TUBE-Based Affinity Enrichment:
- Pre-wash TUBE-conjugated magnetic beads (e.g., LifeSensors UM401M for pan-specific, or linkage-specific equivalents) with lysis buffer.
- Incubate 500-1000 μg of cleared cell lysate with 20-50 μL bead slurry overnight at 4°C with gentle rotation.
- Include parallel samples using different TUBE specificities: pan-specific, K48-specific, and K63-specific TUBEs to enable comparative analysis.
- Wash beads 3-4 times with ice-cold lysis buffer, then elute bound proteins with 2× Laemmli buffer containing 50 mM DTT by heating at 95°C for 5-10 minutes.
Downstream Analysis:
- Analyze eluates by SDS-PAGE and immunoblotting using target-specific antibodies (e.g., anti-RIPK2 for inflammation studies).
- For proteomic applications, digest enriched proteins with trypsin after reduction and alkylation, then analyze by LC-MS/MS.
- To confirm linkage specificity, include controls with linkage-specific TUBEs: K63-TUBEs should enrich L18-MDP-induced RIPK2 ubiquitination, while K48-TUBEs should enrich PROTAC-induced ubiquitination [13].
Protocol for High-Throughput Screening Applications
TUBE-based assays can be adapted to 96-well plate formats for high-throughput screening applications, particularly valuable in drug discovery for characterizing PROTACs and molecular glues [13]:
Plate-Based TUBE Assay:
- Coat 96-well plates with chain-specific TUBEs (K48-TUBE for degradation screening, K63-TUBE for signaling modulation) by adding 100 μL per well of TUBE solution (1-5 μg/mL in PBS) and incubating overnight at 4°C.
- Block plates with 3% BSA in PBS for 2 hours at room temperature, then add cell lysates (prepared as above) and incubate for 3-4 hours at 4°C with gentle shaking.
- Wash plates 4 times with PBS containing 0.05% Tween-20, then detect captured ubiquitinated proteins with target-specific primary antibodies followed by HRP-conjugated secondary antibodies.
- Develop with chemiluminescent substrate and read on a plate luminometer for quantitative assessment.
- Include controls: unstimulated cells, compound-treated cells, and specificity controls with different TUBE types.
Research Reagent Solutions: Essential Materials for TUBE Experiments
Table 3: Essential Research Reagents for TUBE-Based Ubiquitination Studies
| Reagent Category | Specific Examples | Function and Application | Key Features |
|---|---|---|---|
| Pan-Specific TUBEs | LifeSensors UM401M [13] | Global enrichment of ubiquitinated proteins; proteomic studies [28] | High affinity (nM range) for all linkage types; DUB protection [16] |
| K48-Linkage Specific TUBEs | K48-TUBE HF [23] | Selective enrichment of K48-linked chains; degradation studies [13] | ~20 nM affinity for K48 chains; >100-fold selectivity [23] |
| K63-Linkage Specific TUBEs | K63-TUBE [13] | Enrichment of K63-linked chains; signaling studies [13] | Selective for K63 linkages; inflammatory pathway analysis [13] |
| Cell Lines | THP-1 human monocytic cells [13] | Model system for inflammatory signaling studies | NOD2/RIPK2 pathway; responsive to L18-MDP [13] |
| Stimuli & Inhibitors | L18-MDP, Ponatinib, PROTACs [13] | Induce specific ubiquitination; pathway modulation | L18-MDP induces K63 ubiquitination; PROTACs induce K48 ubiquitination [13] |
| DUB Inhibitors | N-ethylmaleimide (NEM) [13] | Preserve ubiquitin chains during lysis | Essential for maintaining endogenous ubiquitination state |
The strategic selection between pan-specific and linkage-specific TUBEs represents a critical decision point in experimental design for ubiquitination research. Pan-specific TUBEs offer comprehensive coverage ideal for discovery-phase research and global ubiquitome profiling, while linkage-specific TUBEs provide functional insights by distinguishing between degradation signals (K48-linked) and non-proteolytic signaling events (K63-linked and other linkages) [13] [23]. The emerging applications of these technologies in high-throughput screening formats, particularly for characterizing PROTACs and molecular glues in drug development, underscore their growing importance in both basic research and therapeutic development [13].
As the ubiquitin field continues to evolve, with increasing recognition of the functional importance of atypical ubiquitin linkages and complex chain architectures, the availability of well-characterized, high-affinity TUBE reagents will remain essential for deciphering the complex language of the ubiquitin code. By applying the decision framework and methodological approaches outlined in this application note, researchers can make informed choices about TUBE selection that align with their specific research objectives, ultimately accelerating progress in understanding ubiquitin-dependent cellular regulation and its therapeutic implications.
Within the ubiquitin-proteasome system, the native ubiquitination state of a protein is a fleeting snapshot of cellular activity, capturing critical information about protein fate, from degradation signaling to immune response regulation. For researchers using Tandem-repeated Ubiquitin-Binding Entities (TUBEs) to study these processes, the integrity of this snapshot is paramount. The very first step in the workflow—cell lysis and sample preparation—is the most critical point where this delicate information can be lost. The reversible nature of ubiquitination, driven by active deubiquitylases (DUBs), means that standard lysis protocols are often inadequate, leading to the rapid erasure of ubiquitin signals before analysis can begin. This application note details optimized lysis and preparation protocols designed specifically to preserve the native ubiquitylation state of proteins, ensuring that subsequent enrichment with TUBEs provides a biologically relevant and accurate readout for drug development professionals and research scientists.
The Critical Importance of Preserving Ubiquitylation
Protein ubiquitylation is a highly dynamic and reversible post-translational modification. During cell lysis, the disruption of cellular compartments releases active DUBs, which can rapidly hydrolyze ubiquitin chains from substrate proteins. Furthermore, the 26S proteasome continuously degrades proteins marked with certain ubiquitin chains, primarily K48-linked polymers. Therefore, to accurately capture the endogenous state of protein ubiquitylation—a necessity for valid TUBE-based enrichment—these processes must be effectively inhibited at the moment of lysis.
The challenge is twofold: first, to instantaneously inhibit DUB activity to prevent deubiquitylation, and second, to stabilize the ubiquitin-protein conjugates long enough for them to be captured. Failure to do so results in the underestimation of ubiquitylation levels, potential misinterpretation of the predominant ubiquitin chain linkage, and a failure to detect transient but biologically critical ubiquitylation events, such as those induced by PROTACs or inflammatory stimuli [29] [13].
Optimized Strategies for Lysis and Preservation
Inhibition of Deubiquitylases (DUBs)
DUBs are predominantly cysteine proteases, with one family encoding metalloproteinases. Effective inhibition requires a combination of agents to target these different enzyme classes.
- Cysteine Protease Inhibitors: N-Ethylmaleimide (NEM) and Iodoacetamide (IAA) are alkylating agents that modify the active site cysteine residue of DUBs. While concentrations of 5-10 mM are commonly reported, research indicates that up to 10-fold higher concentrations (50-100 mM) may be required to fully preserve the ubiquitylation status of some substrates, such as IRAK1 [29]. NEM is generally more effective than IAA at preserving K63- and M1-linked ubiquitin chains and is the preferred agent when the downstream analysis includes mass spectrometry, as its adducts do not interfere with the identification of ubiquitylation sites [29].
- Metalloprotease Inhibitors: Chelating agents such as EDTA or EGTA (typically at 1-10 mM) must be included in the lysis buffer to sequester heavy metal ions required by the metalloprotease family of DUBs [29] [30].
- Broad-Spectrum DUB Inhibitors: Compounds like PR-619 provide a broader inhibition profile and can be used in conjunction with the agents above for more comprehensive protection [31].
Table 1: Common DUB Inhibitors for Lysis Buffers
| Inhibitor | Typical Working Concentration | Mechanism of Action | Key Considerations |
|---|---|---|---|
| N-Ethylmaleimide (NEM) | 10 - 100 mM | Alkylates active site cysteine residues | Preferred for MS; more stable than IAA; better for K63/M1 chains [29] |
| Iodoacetamide (IAA) | 10 - 100 mM | Alkylates active site cysteine residues | Degrades rapidly in light; its adduct can interfere with MS-based ubiquitylation site mapping [29] |
| EDTA/EGTA | 1 - 10 mM | Chelates metal ions (Zn²⁺) | Inhibits metallo-DUBs [29] |
| PR-619 | 10 - 50 µM | Reversible, cell-permeable DUB inhibitor | Used as a broad-spectrum inhibitor in some specialized protocols [31] |
Proteasome Inhibition
To prevent the loss of polyubiquitylated proteins targeted for degradation, inhibition of the proteasome is essential. This is particularly important for studying K48-linked ubiquitylation.
- MG132: This cell-permeable tripeptide aldehyde is a potent and reversible inhibitor of the proteasome's chymotrypsin-like activity. Treating cells with 10-25 µM MG132 for 2-6 hours prior to lysis allows for the accumulation and stabilization of proteasome-targeted ubiquitylated proteins, such as pUb-IκBα, facilitating their detection [29] [32].
- Considerations: Prolonged treatment with MG132 (e.g., 12-24 hours) can induce cytotoxic stress responses, which may themselves alter the cellular ubiquitylation landscape. Therefore, incubation times should be carefully optimized for each cell system [29].
Lysis Buffer Composition
The choice of lysis buffer is critical for inactivating enzymes, extracting the protein of interest, and maintaining protein interactions. Strong denaturing conditions offer the best protection for ubiquitin conjugates.
- Strong Denaturing Lysis: Directly lysing cells in a buffer containing 1% SDS and immediate boiling provides the most effective and instantaneous inactivation of DUBs and other enzymes. This method is ideal when the sole objective is to analyze the ubiquitylation state by immunoblotting [29].
- Urea-Based Lysis: For workflows that require subsequent incubations, such as TUBE pull-downs or immunoprecipitation, a lysis buffer containing 6-8 M urea or 4-6 M guanidine hydrochloride offers a strong denaturing environment that preserves ubiquitylation while keeping proteins in a solution compatible with these procedures [30] [31].
- Compatible (Mild) Lysis: If maintaining protein-protein interactions or enzyme activity is necessary, a non-denaturing lysis buffer (e.g., containing 1% Triton X-100 or NP-40) can be used, but only with a potent cocktail of DUB and protease inhibitors, as described above [13] [33].
Table 2: Lysis Buffer Formulations for Ubiquitylation Studies
| Buffer Type | Example Composition | Best Use Case | Advantages & Disadvantages |
|---|---|---|---|
| Strong Denaturing | 1% SDS, 50 mM Tris pH 8.0, 150 mM NaCl, + DUB inhibitors [29] | Direct immunoblot analysis of ubiquitylation | Best DUB inhibition Incompatible with IP/TUBE pull-downs |
| Urea-Based Denaturing | 8 M Urea, 50 mM Tris pH 8.0, 150 mM NaCl, 1 mM EDTA, + DUB inhibitors [31] | TUBE enrichment; MS sample prep | Strong denaturation, TUBE compatible Requires desalting for some downstream steps |
| Mild Non-Denaturing | 1% Triton X-100, 50 mM Tris pH 8.0, 150 mM NaCl, 10% Glycerol, + DUB/protease inhibitors [13] [33] | Studying ubiquitin-mediated protein complexes | Preserves native interactions Higher risk of deubiquitylation |
Step-by-Step Protocol for Optimized Sample Preparation
Materials and Reagents
- Proteasome Inhibitor: MG132 (e.g., Selleckchem S2619)
- DUB Inhibitors: NEM (Sigma-Aldrich E3876), EDTA (Sigma-Aldrich E7889)
- Lysis Buffer: Choose from formulations in Table 2.
- Protease Inhibitor Cocktail: Commercial tablet (e.g., Roche 05892791001) or custom mix (e.g., 1 mM PMSF, 5 µg/mL Leupeptin, 2 µg/mL Aprotinin) [30]
- Phosphate-Buffered Saline (PBS)
- Cell Scraper (for adherent cells)
- Microcentrifuge (capable of 14,000-16,000 g)
- Sonicator (probe or bath)
Pre-Lysis: Cell Treatment and Harvesting
- Proteasome Inhibition: Approximately 2-4 hours before harvesting, add MG132 to the cell culture medium to a final concentration of 10-25 µM. Return cells to the incubator [29] [32].
- Harvesting:
- For adherent cells: Aspirate the medium and wash cells gently with ice-cold PBS. Aspirate PBS completely.
- For suspension cells: Pellet cells by centrifugation (e.g., 300 g for 5 min at 4°C). Aspirate supernatant and wash pellet with ice-cold PBS.
Lysis Procedure
This protocol describes the urea-based lysis method, which is broadly applicable for TUBE-based workflows.
- Prepare Lysis Buffer Fresh: Just before use, add DUB and protease inhibitors to the urea lysis buffer. For example, to 10 mL of urea lysis buffer, add:
- 100 µL of 1 M NEM (Final: 10 mM)
- 20 µL of 0.5 M EDTA (Final: 1 mM)
- One tablet of protease inhibitor cocktail.
- Lyse Cells:
- For adherent cells: Add an appropriate volume of lysis buffer directly to the culture dish (e.g., 150 µL for a 35-mm dish). Rock the dish to ensure complete coverage. Scrape the cells and transfer the lysate to a pre-cooled microcentrifuge tube.
- For suspension cells: Resuspend the cell pellet in an appropriate volume of lysis buffer by gentle pipetting.
- Vortex the tubes briefly and place on ice for 10-15 minutes.
- Sonicate: To reduce viscosity and shear genomic DNA, sonicate the lysates on ice. Use a microtip sonicator with short bursts (e.g., 3 pulses of 5 seconds each at 20% amplitude), allowing the samples to cool on ice between pulses. Alternatively, a bath sonicator can be used.
- Clarify Lysate: Centrifuge the lysates at 14,000-16,000 g for 15 minutes at 4°C to pellet insoluble material.
- Collect Supernatant: Carefully transfer the clarified supernatant (the protein lysate) to a new, pre-cooled microcentrifuge tube.
- Protein Quantification: Determine the protein concentration using a compatible assay (e.g., BCA assay). The lysate can now be used immediately for TUBE-based enrichment or frozen at -80°C for future use.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for Ubiquitylation Preservation and Analysis
| Reagent / Tool | Function | Example Product / Assay |
|---|---|---|
| TUBEs (Pan-specific) | High-affinity capture of all ubiquitin chain linkages; protects chains from DUBs during enrichment [29] [34] | LifeSensors TUBE Kits (e.g., UM401M) [13] [34] |
| Chain-Selective TUBEs | Selective enrichment of specific ubiquitin linkages (e.g., K48, K63) to decipher chain topology [13] [16] | LifeSensors K48-TUBE, K63-TUBE [13] |
| DUB Inhibitors (NEM, PR-619) | Preserve ubiquitin conjugates by alkylating active site cysteines of deubiquitylases during lysis [29] [31] | Sigma-Aldrich E3876 (NEM); SML0430 (PR-619) |
| Proteasome Inhibitor (MG132) | Stabilizes K48-polyubiquitylated proteins by blocking their degradation by the 26S proteasome [29] [32] | Selleckchem S2619 (MG132) |
| Ubiquitin-Trap | Nanobody-based reagent for immunoprecipitation of mono-Ub, poly-Ub, and ubiquitylated proteins [32] | ChromoTek Ubiquitin-Trap Agarose (uta) |
| Linkage-Specific Antibodies | Detect specific ubiquitin chain types (K48, K63, M1, etc.) via immunoblotting after enrichment [16] | Cell Signaling Technology, Proteintech |
Workflow Visualization
Ubiquitin Signal Preservation and Analysis Workflow
The following diagram illustrates the critical steps for preserving and analyzing protein ubiquitylation, from live cell to final detection:
DUB Inhibition Mechanism
The mechanism by which DUB inhibitors prevent the loss of ubiquitin signals during sample preparation is detailed below:
The successful enrichment and analysis of ubiquitinated proteins using TUBEs is fundamentally dependent on the initial steps of sample preparation. By implementing a lysis strategy that emphasizes rapid and potent inhibition of DUBs and the proteasome, researchers can effectively "freeze" the native ubiquitination state of the cellular proteome. The protocols and guidelines provided here, centered on the use of high-concentration alkylating agents like NEM and strong denaturing buffers, form a robust foundation for obtaining reliable, high-quality data that accurately reflects the biological state of the ubiquitin system. This rigorous approach to sample integrity is essential for advancing research in targeted protein degradation, inflammatory signaling, and the development of novel therapeutics that modulate the ubiquitin pathway.
Step-by-Step Guide to TUBE-Based Immunoprecipitation and Pull-Down Assays
The study of protein ubiquitination has been revolutionized by the development of Tandem Ubiquitin Binding Entities (TUBEs), engineered protein domains that specifically bind to polyubiquitin chains with nanomolar affinity [5] [6]. Unlike traditional methods that rely on ubiquitin antibodies—which are often notoriously non-selective and can lead to artifacts—TUBEs provide a highly sensitive and specific means to isolate polyubiquitylated proteins from complex cell lysates and tissues [6] [16]. The versatility of ubiquitination, which can form chains of different lengths and linkage types, regulates diverse cellular functions including protein degradation, signal transduction, and DNA repair [16] [13]. Understanding these specific ubiquitin signals is crucial for advancing drug discovery, particularly in the field of Targeted Protein Degradation (TPD) with PROTACs (Proteolysis Targeting Chimeras) and molecular glues [5] [13].
A key advantage of TUBEs is their ability to protect ubiquitylated proteins from both deubiquitinating enzymes (DUBs) and proteasome-mediated degradation, even in the absence of the inhibitors normally required to block these activities [6]. This protective function preserves the native ubiquitination state of proteins during experimental procedures. TUBEs exist in two primary forms: pan-selective TUBEs that bind to all types of polyubiquitin chains, and chain-selective TUBEs that specifically recognize particular linkage types such as K48, K63, or M1 (linear) chains [5] [6]. This protocol article provides a comprehensive guide to implementing TUBE-based assays for the enrichment and study of ubiquitinated proteins, framed within the broader context of ubiquitinated protein enrichment research.
Principles and Advantages of TUBE-Based Assays
Theoretical Foundations of TUBE Technology
TUBE technology harnesses the strength of multiple Ubiquitin Binding Domains (UBDs) arranged in tandem, creating an affinity matrix with significantly enhanced binding capability compared to single UBDs [6] [16]. This arrangement overcomes the low affinity limitations of individual UBDs, enabling efficient capture of polyubiquitinated proteins without requiring overexpression of epitope-tagged ubiquitin [6]. The fundamental principle relies on the specific interaction between these engineered tandem domains and the polyubiquitin chains attached to target proteins, with dissociation constants (Kd) typically in the low nanomolar range (1-10 nM) [6].
The complexity of ubiquitin signaling arises from the ability of ubiquitin to form polymers through eight different linkage sites (M1, K6, K11, K27, K29, K33, K48, K63), each potentially conferring distinct functional consequences [16] [13]. For example, K48-linked chains primarily target substrates for proteasomal degradation, while K63-linked chains regulate signal transduction and protein trafficking [13]. TUBE technology addresses this complexity through linkage-specific variants that can discriminate between these different chain types, enabling researchers to investigate the specific ubiquitin code associated with cellular processes or therapeutic interventions.
Comparative Advantages Over Traditional Methods
Table: Comparison of Ubiquitin Enrichment Methods
| Method | Sensitivity | Specificity | Ability to Preserve Ubiquitination | Throughput Potential | Cost Considerations |
|---|---|---|---|---|---|
| TUBEs | High (nanomolar affinity) | High (especially chain-selective TUBEs) | Excellent (protects from DUBs/proteasomes) | High (adaptable to HTS formats) | Moderate |
| Ub Antibodies | Variable | Often non-selective [6] | Limited (requires inhibitors) | Low to moderate | High (especially large-scale studies) |
| Tagged Ubiquitin | Moderate | High for tagged proteins | Limited to overexpression systems | Moderate | Low to moderate |
| Mass Spectrometry without Enrichment | Low for ubiquitinated species | High when sites identified | N/A | Low | High |
Traditional immunoprecipitation using ubiquitin antibodies faces limitations including high cost for large-scale studies, variable specificity, and inability to protect ubiquitin chains from deubiquitination during processing [6] [16]. The tagged ubiquitin approach (e.g., His- or Strep-tagged Ub) requires genetic manipulation of cellular systems and may not accurately represent endogenous ubiquitination patterns [16]. TUBEs overcome these limitations by providing a versatile tool that preserves native ubiquitination states, functions with endogenous protein levels, and offers specificity for chain types when needed [6] [13].
Essential Reagents and Materials
The Scientist's Toolkit: Research Reagent Solutions
Table: Essential Reagents for TUBE-Based Assays
| Reagent/Material | Function/Purpose | Examples/Specifications |
|---|---|---|
| TUBE Reagents | Specific capture of polyubiquitinated proteins | Pan-selective (UM401M [13]), K48-TUBE, K63-TUBE, M1-TUBE [6] |
| Lysis Buffer | Cell disruption while preserving protein interactions | NP-40 buffer (mild): 150 mM NaCl, 1% NP-40, 50 mM Tris-HCl pH=8.0 [35] |
| Protease Inhibitors | Prevent protein degradation during processing | Commercial cocktails (e.g., ab65621) [35] |
| Phosphatase Inhibitors | Preserve phosphorylation states | Added when studying phosphorylated proteins [35] |
| Magnetic Beads | Solid support for TUBE immobilization | TUBE1-conjugated magnetic beads (UM401M) [13] |
| DUB Inhibitors | Optional additional protection against deubiquitination | N-ethylmaleimide or specific DUB inhibitors |
| Wash Buffers | Remove non-specifically bound proteins | Varied stringency (e.g., low to moderate salt) |
| Elution Buffers | Release captured ubiquitinated proteins | SDS-PAGE sample buffer or competitive elution with free ubiquitin |
Successful TUBE-based assays require careful selection of appropriate TUBE types based on experimental goals. For general profiling of ubiquitinated proteins, pan-selective TUBEs such as TUBE1 (UM401M from LifeSensors) are ideal [13]. When investigating specific functional outcomes—such as proteasomal targeting (K48-linkages) or signaling events (K63-linkages)—chain-selective TUBEs provide the necessary specificity [6] [13]. The choice of lysis buffer is critical; non-ionic detergents like NP-40 are less harsh than ionic detergents and help maintain protein interactions [35]. Consistently including protease inhibitors in all buffers is essential to prevent degradation of ubiquitin chains and target proteins during the enrichment process.
Diagram 1: TUBE selection decision tree. This flowchart guides researchers in selecting the appropriate TUBE type based on their specific experimental questions and the biological processes under investigation.
Detailed Experimental Protocols
Stage 1: Cell Lysis and Sample Preparation
Principles: Effective cell lysis must balance complete disruption of cellular structures with preservation of protein interactions and ubiquitination states. The selection of lysis buffer depends on protein localization and experimental requirements [35].
Step-by-Step Protocol:
- Prepare ice-cold lysis buffer: Use NP-40 lysis buffer (150 mM NaCl, 1% NP-40, 50 mM Tris-HCl pH=8.0) for cytoplasmic/membrane proteins, or RIPA buffer (50 mM Tris-HCl pH=8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) for nuclear proteins or more stringent conditions [35].
- Add inhibitors: Supplement buffer with protease inhibitor cocktail immediately before use. Include phosphatase inhibitors if studying phosphorylated proteins [35].
- Harvest cells: Wash cells with PBS and pellet by centrifugation. For tissue samples, snap-freeze in liquid nitrogen and homogenize using a bead beater homogenizer with appropriate lysis buffer [35].
- Lyse cells: Resuspend cell pellet in ice-cold lysis buffer (300 µL for 1-3×10^7 cells). Incubate on ice for 10 minutes without agitation [35].
- Clarify lysate: Centrifuge at 8,000 × g for 10 minutes at 4°C. Transfer supernatant to a fresh tube kept on ice [35].
- Determine protein concentration: Use Bradford or BCA assay. Adjust concentrations if necessary, or consider fractionation for low-abundance proteins [35].
- Aliquot and store: Snap-freeze aliquots in liquid nitrogen and store at -80°C if not used immediately.
Critical Notes:
- Maintain samples, buffers, and equipment on ice throughout the process.
- Avoid repeated freeze-thaw cycles of lysates.
- Optimize centrifugation force and time for specific cell types (e.g., leukocytes require lighter centrifugation) [35].
Stage 2: TUBE-Based Pull-Down Assay
Principles: This stage utilizes the high affinity of TUBEs for polyubiquitin chains to selectively enrich ubiquitinated proteins from the prepared lysates. The choice between pan-selective and chain-selective TUBEs depends on experimental goals [5] [6].
Step-by-Step Protocol:
- Pre-clear lysate (optional): Incubate lysate with bare beads or beads plus isotype control antibody to reduce non-specific binding. This step may be skipped when using high-quality beads without significant bead adsorption effects [35].
- Prepare TUBE-bead complex: If using unconjugated TUBEs, immobilize onto appropriate beads (e.g., magnetic beads, agarose) according to manufacturer's instructions. For pre-conjugated TUBE beads (e.g., UM401M), wash with lysis buffer before use [13].
- Incubate lysate with TUBE beads: Add 500 µg–1 mg of total protein lysate to prepared TUBE beads. Adjust volume with lysis buffer to maintain consistent binding conditions.
- Binding reaction: Incubate with gentle rotation for 2–4 hours at 4°C to maximize binding while minimizing degradation.
- Wash beads: Collect beads (via magnetization for magnetic beads or centrifugation for agarose) and wash 3–4 times with appropriate wash buffer (e.g., lysis buffer with 300–500 mM NaCl for higher stringency).
- Elute bound proteins: Elute ubiquitinated proteins by adding 2× SDS-PAGE sample buffer and heating at 95°C for 5–10 minutes. Alternatively, competitive elution can be performed using free ubiquitin (3–5 mg/mL).
Critical Notes:
- Include appropriate controls: (1) beads alone, (2) isotype control (for antibody-based detection), and (3) non-specific competitor (free ubiquitin).
- For chain-specific studies, validate specificity with appropriate controls (e.g., known K48- vs K63-ubiquitinated proteins) [13].
- Keep incubation times consistent across samples for comparative studies.
Stage 3: Downstream Analysis and Applications
Principles: The eluted proteins can be analyzed by various methods depending on research questions. Western blotting provides information about specific targets, while mass spectrometry enables global profiling of ubiquitinated proteins [35] [16].
Western Blot Analysis:
- Separate eluted proteins by SDS-PAGE appropriate for target protein size.
- Transfer to PVDF membrane for immunoblotting.
- Probe with antibodies against protein(s) of interest to detect ubiquitinated forms (typically appearing as higher molecular weight smears).
- For confirmation of ubiquitination, reprobe with ubiquitin-specific antibodies.
- Use chain-specific ubiquitin antibodies to determine linkage types when combined with chain-selective TUBE pull-downs.
Mass Spectrometry Analysis:
- Process eluted proteins for MS analysis (reduction, alkylation, digestion).
- For ubiquitination site mapping, use tryptic digestion which generates di-glycine remnants on modified lysines (mass shift of 114.04 Da) [16].
- Analyze peptides by LC-MS/MS to identify ubiquitination sites and relative abundance.
- For comprehensive ubiquitome analysis, combine TUBE enrichment with quantitative proteomics (e.g., SILAC, TMT).
High-Throughput Applications:
- Plate-based assays: Immobilize TUBEs on microtiter plates for higher throughput analysis of ubiquitination in response to drugs, inhibitors, or PROTAC molecules [5].
- PROTAC characterization: Use TUBE-based assays to monitor target protein ubiquitination induced by PROTAC treatment, enabling rank-order potency assessment [13].
- Drug screening: Implement TUBE-based platforms to identify compounds that modulate ubiquitination of specific targets.
Diagram 2: TUBE assay workflow and applications. This visualization outlines the key stages of TUBE-based pull-down assays and the primary downstream analysis methods, highlighting the connectivity between experimental phases and their applications in ubiquitination research.
Data Analysis and Interpretation
Quantitative Assessment and Normalization
Western Blot Quantification:
- Quantify ubiquitination signals using densitometry analysis software.
- Normalize ubiquitination signals to total input protein or housekeeping genes.
- For time-course experiments, express data as fold-change relative to control conditions.
- Present ubiquitinated proteins as higher molecular weight species or smears characteristic of polyubiquitination.
Mass Spectrometry Data Analysis:
- Identify ubiquitination sites through detection of di-glycine remnant (K-ε-GG) peptides.
- Apply appropriate false discovery rate (FDR) thresholds (typically <1%).
- Use spectral counting or label-free quantification for relative abundance measurements.
- Perform pathway enrichment analysis using databases like KEGG or Gene Ontology to identify biological processes regulated by ubiquitination [36].
Troubleshooting Common Issues
Table: Troubleshooting Guide for TUBE-Based Assays
| Problem | Potential Causes | Solutions |
|---|---|---|
| High background | Non-specific binding | Increase wash stringency (higher salt), include pre-clearing step, optimize TUBE:lysate ratio |
| Low signal | Insufficient ubiquitinated proteins | Increase input protein, check protease inhibitor efficacy, verify cell stimulation conditions |
| Incomplete elution | Strong TUBE-ubiquitin interaction | Use more denaturing elution conditions, increase elution temperature/time |
| Inconsistent results | Variable binding conditions | Standardize incubation times, maintain consistent temperature, use fresh inhibitors |
| No chain specificity | Cross-reactivity of TUBEs | Validate TUBE specificity with known standards, check manufacturer's recommendations |
Application Case Study: RIPK2 Ubiquitination Analysis
A recent study demonstrated the power of chain-selective TUBEs in investigating context-dependent ubiquitination of RIPK2, a key regulator of inflammatory signaling [13]. Researchers used THP-1 cells treated with either L18-MDP (to induce inflammatory signaling) or a RIPK2-targeting PROTAC (to induce degradation).
Experimental Design:
- Stimulation: THP-1 cells were treated with L18-MDP (200-500 ng/ml) for 30-60 minutes to induce K63-linked ubiquitination or with RIPK2 PROTAC to induce K48-linked ubiquitination [13].
- Inhibition: Some experiments included pre-treatment with Ponatinib (100 nM), a RIPK2 inhibitor, for 30 minutes before stimulation [13].
- Lysis: Cells were lysed in buffer optimized to preserve polyubiquitination.
- TUBE Pull-Down: Lysates were incubated with pan-selective, K48-selective, or K63-selective TUBEs.
- Analysis: Captured proteins were analyzed by immunoblotting with anti-RIPK2 antibody.
Key Findings:
- L18-MDP stimulation induced K63 ubiquitination of RIPK2, captured efficiently by K63-TUBEs and pan-TUBEs but not K48-TUBEs.
- RIPK2 PROTAC induced K48 ubiquitination, captured by K48-TUBEs and pan-TUBEs but not K63-TUBEs.
- Ponatinib treatment completely abrogated L18-MDP-induced RIPK2 ubiquitination [13].
This case study highlights how chain-selective TUBEs can discriminate between different functional ubiquitin linkages on the same protein in different cellular contexts, providing crucial insights for targeted drug development.
TUBE-based immunoprecipitation and pull-down assays represent a significant advancement in ubiquitination research, addressing critical limitations of traditional methods. The high affinity, specificity, and protective capabilities of TUBEs make them invaluable tools for studying the complex landscape of protein ubiquitination. As research in targeted protein degradation continues to expand, particularly in the development of PROTACs and molecular glues, TUBE technology provides a robust platform for characterizing compound efficacy, mechanism of action, and specificity. By following the detailed protocols outlined in this guide, researchers can reliably implement these methods to advance our understanding of ubiquitin-dependent processes in health and disease.
Protein ubiquitination is a crucial post-translational modification that influences nearly all cellular processes, including proteasomal degradation, cell signaling, DNA repair, and immune responses [13]. The functional outcome of ubiquitination is determined by the type of polyubiquitin chain formed on the substrate protein. Among the eight distinct chain linkages, K48-linked chains primarily target proteins for proteasomal degradation, while K63-linked chains are largely involved in non-proteolytic signaling pathways, such as inflammation and autophagy [13] [37]. Unraveling this "ubiquitin code" is fundamental to understanding cellular biology and developing new therapeutics, but it remains analytically challenging due to the low abundance of ubiquitinated proteins, the activity of deubiquitinases (DUBs), and the transient nature of the modifications.
Tandem Ubiquitin Binding Entities (TUBEs) are engineered, high-affinity reagents designed to overcome these challenges. Composed of multiple ubiquitin-associated (UBA) domains, TUBEs bind polyubiquitin chains with nanomolar affinity, shielding them from deubiquitinating enzymes and proteasomal degradation during isolation [13] [38] [37]. This application note details how chain-selective and pan-selective TUBEs are revolutionizing proteomics, E3 ligase substrate identification, and high-throughput drug discovery.
TUBE Technology: A Research Reagent Solution
TUBEs are versatile tools available in various formats tailored for specific research applications. The table below summarizes the key reagent solutions and their primary functions.
Table 1: Key Research Reagent Solutions Using TUBE Technology
| Reagent Solution | Core Function | Key Features and Applications |
|---|---|---|
| Pan-Selective TUBEs (e.g., TUBE1, TUBE2) | Broad capture of all ubiquitin chain linkage types (K6, K11, K27, K29, K33, K48, K63, M1) [37]. | Ideal for general ubiquitome studies, protecting polyubiquitinated proteins from DUBs, and total ubiquitin pull-down for mass spectrometry [38] [37]. |
| K48-Selective TUBEs | Specifically enrich for K48-linked polyubiquitin chains [37]. | Powerful tool for studying proteasomal degradation pathways and validating PROTAC activity [13] [34]. |
| K63-Selective TUBEs | Exhibit a 1,000 to 10,000-fold preference for K63-linked chains [37]. | Used to investigate autophagy, lysosomal degradation, DNA repair, and inflammatory signaling (e.g., NF-κB, NLRP3 inflammasome) [13] [37]. |
| M1-Linear Selective TUBEs | Target linear (M1-linked) ubiquitin chains [34]. | Applied to identify degraders of targets in inflammatory pathways [34]. |
| TUBE-Embedded Microtiter Plates | High-throughput plate-based assays for ubiquitination detection [34]. | Enable screening for molecular glues and PROTACs in a live-cell, high-throughput format without tags or artificial modifications [19] [34]. |
| Phospho-TUBEs (In development) | Designed to isolate phosphorylated ubiquitin chains (e.g., Ser65-phosphorylated Ub) [37]. | A cutting-edge tool for studying mitophagy and neurodegenerative diseases like Parkinson's disease, where PINK1 phosphorylates ubiquitin [37]. |
Application Note 1: Chain-Specific Ubiquitination Analysis in Drug Discovery
Background and Protocol
Targeted protein degradation (TPD) via Proteolysis Targeting Chimeras (PROTACs) and molecular glues is a transformative therapeutic strategy. A critical challenge in this field is the high-throughput assessment of whether a candidate molecule induces the intended ubiquitination of the target protein and does so with the correct linkage (typically K48 for degradation) [13]. Traditional methods like Western blotting are low-throughput and semi-quantitative, while mass spectrometry is labor-intensive.
A 2025 study demonstrated the use of chain-specific TUBEs in HTS assays to investigate the ubiquitination dynamics of RIPK2, a key regulator of inflammatory signaling [13]. The protocol below outlines this application.
Table 2: Protocol for Chain-Specific Analysis of RIPK2 Ubiquitination using TUBEs
| Step | Procedure | Critical Parameters |
|---|---|---|
| 1. Cell Stimulation | Treat human monocytic THP-1 cells with:• Inflammatory stimulus: L18-MDP (200-500 ng/ml, 30-60 min) to induce K63-linked ubiquitination.• Degradation stimulus: RIPK2 PROTAC (e.g., RIPK degrader-2) to induce K48-linked ubiquitination [13]. | Include controls (vehicle) and pre-treatment with specific inhibitors (e.g., Ponatinib for RIPK2) to validate signal specificity [13]. |
| 2. Cell Lysis | Lyse cells in a buffer optimized to preserve polyubiquitination. | The lysis buffer must contain DUB inhibitors and be compatible with downstream TUBE binding [13]. |
| 3. TUBE-Based Enrichment | Incubate cell lysates with 96-well plates coated with Pan-, K48-, or K63-selective TUBEs [13]. | Use equivalent amounts of total protein across conditions. The high affinity of TUBEs allows for stringent washing to reduce background. |
| 4. Detection & Analysis | Detect captured ubiquitinated RIPK2 using an anti-RIPK2 antibody via plate-based readout (e.g., luminescence) [13]. | The HTS-compatible format allows for quantitative, multi-well analysis of endogenous protein ubiquitination without transfection or tagging. |
Key Findings and Data Interpretation
The application of this protocol yielded clear, linkage-specific results:
- L18-MDP stimulation induced RIPK2 ubiquitination that was captured by K63-TUBEs and Pan-TUBEs, but not by K48-TUBEs [13].
- Conversely, RIPK2 PROTAC treatment induced ubiquitination captured by K48-TUBEs and Pan-TUBEs, with no appreciable signal in the K63-TUBE assay [13].
- The RIPK2 inhibitor Ponatinib completely abrogated L18-MDP-induced ubiquitination, confirming the specificity of the signal [13].
This demonstrates that chain-selective TUBEs can effectively differentiate context-dependent ubiquitin linkages on an endogenous target protein in a high-throughput format, providing a powerful method for characterizing PROTACs and molecular glues.
Diagram 1: RIPK2 ubiquitination pathways. Inflammatory and degradation stimuli trigger distinct ubiquitin linkages and cellular outcomes, which can be differentiated using chain-specific TUBEs.
Application Note 2: Proteomic Identification of Ubiquitin Ligase Substrates with BioE3
Background and Protocol
Identifying the specific substrates for the ~600 human E3 ubiquitin ligases is a monumental challenge in cell biology, as it requires distinguishing genuine ubiquitination targets from mere interactors. The BioE3 method is an innovative strategy that combines TUBE-like affinity with proximity-dependent biotinylation to label and isolate bona fide substrates of specific E3 ligases [39].
This method involves creating a fusion protein between the E3 ligase of interest and BirA, a bacterial biotin ligase. This is combined with a "bioGEFUb" construct—a ubiquitin molecule fused to a modified AviTag (bioGEF) that has lower affinity for BirA, which minimizes non-specific labeling. When the BirA-E3 fusion ubiquitinates a substrate with the bioGEFUb, the proximity allows BirA to biotinylate the bioGEF tag. This covalent biotin mark enables stringent streptavidin-based purification of the substrate for identification by mass spectrometry [39].
Table 3: Protocol for E3 Ligase Substrate Identification using BioE3
| Step | Procedure | Critical Parameters |
|---|---|---|
| 1. Cell Line Engineering | Generate stable cell lines (e.g., HEK293FT, U2OS) with inducible expression of bioGEFUb (wild-type or non-cleavable mutant) [39]. | Using the bioGEF tag, not the wild-type AviTag (bioWHE), is crucial to avoid non-specific, proximity-independent biotinylation [39]. |
| 2. BirA-E3 Expression | Introduce the BirA-E3 ligase fusion construct into the bioGEFUb cells. | Fuse BirA to the N-terminus of RING-type E3s to avoid steric interference with the C-terminal RING domain [39]. |
| 3. Biotin Depletion & Labeling | Culture cells in biotin-depleted media. Induce bioGEFUb and BirA-E3 expression with doxycycline. Add exogenous biotin for a limited time (e.g., 2 hours) [39]. | Controlling biotin availability and timing is essential for limiting biotinylation to true, proximity-dependent events during the ubiquitination reaction. |
| 4. Affinity Purification & MS | Lyse cells and perform streptavidin-based pull-down under denaturing conditions. Identify captured proteins via Liquid Chromatography-Mass Spectrometry (LC-MS) [39]. | Stringent washing is required. Identified proteins are high-confidence substrates of the E3 ligase of interest. |
Key Findings and Data Interpretation
The BioE3 method has been successfully validated and applied to multiple E3 ligases:
- Validation: BioE3 correctly identified known substrates of well-characterized E3 ligases like RNF4 (in DNA damage) and MIB1 (in endocytosis and centrosome dynamics) [39].
- Versatility: The method works for different types of E3s, including RING-type ligases (e.g., mitochondrial MARCH5, uncharacterized RNF214) and HECT-type ligases (e.g., NEDD4) [39].
- Specificity: The use of bioGEFUb virtually eliminated the non-specific background labeling that was pervasive with the original bioWHEUb tag, confirming the importance of the engineered tag for specificity [39].
This technology enables the creation of a much-needed "compendium of targets for specific E3 ligases," shedding light on the complex ubiquitin network and opening new avenues for drug discovery by identifying novel, disease-relevant substrates [39].
Diagram 2: BioE3 workflow for substrate ID. The method uses a BirA-E3 fusion and bioGEFUb to label and isolate true E3 substrates for mass spectrometry.
Application Note 3: TUBEs in Mass Spectrometry Proteomics
Background and Protocol
Mass spectrometry (MS)-based proteomics is a powerful, untargeted approach for discovering ubiquitinated proteins across the proteome. However, the low stoichiometry of ubiquitination and the susceptibility of ubiquitin chains to DUBs make this analysis difficult. TUBEs serve as an ideal affinity matrix for MS sample preparation by enabling efficient enrichment of polyubiquitinated proteins while preserving their native state [38].
Pan-selective TUBEs capture all ubiquitin chain linkage types, providing a comprehensive view of the ubiquitome and revealing dynamic ubiquitin remodeling in different cellular contexts [38]. Furthermore, using chain-selective TUBEs (e.g., K48- or K63-specific) allows for a deeper, more precise exploration, enabling researchers to determine the specific types of ubiquitination a protein or system undergoes [38]. This is invaluable for understanding functional consequences.
Table 4: Protocol for Ubiquitin Proteomics using TUBE-based Enrichment
| Step | Procedure | Critical Parameters |
|---|---|---|
| 1. Sample Preparation | Lyse cells or tissues in a denaturing buffer (e.g., containing SDS) to inactivate DUBs and preserve the ubiquitination state. | Rapid processing and the use of DUB inhibitors in the initial lysis step are critical to prevent artifactually altered ubiquitin profiles [38]. |
| 2. TUBE Pull-Down | Dilute the lysate to a detergent concentration compatible with TUBE binding. Incubate with Pan- or Chain-selective TUBE magnetic beads. | The high affinity of TUBEs allows for stringent washing to reduce non-specific binding, improving the signal-to-noise ratio for MS. |
| 3. On-Bead Digestion | Wash the beads and subject the captured proteins to on-bead tryptic digestion to generate peptides for LC-MS/MS. | This step can be performed using commercial kits, such as the LifeSensors Ubiquitin Mass Spectrometry Kit (UM420) [38]. |
| 4. LC-MS/MS & Data Analysis | Analyze the resulting peptides by Liquid Chromatography-Tandem Mass Spectrometry. | Data analysis can identify ubiquitinated proteins and, depending on the method, potentially map specific ubiquitination sites. |
Key Findings and Data Interpretation
TUBE-based MS proteomics offers several key advantages over other methods:
- Overcoming Antibody Bias: Pan-selective TUBEs capture all ubiquitin chain linkages, overcoming the inherent bias of antibody-based methods like di-glycine (diGly) remnant enrichment, which may have variable efficiency for different chain types [38].
- Preservation of Native Architecture: By shielding ubiquitin chains from DUBs, TUBEs allow for the analysis of intact polyubiquitinated proteins, providing insights into chain topology that are lost in peptide-centric diGly approaches [38].
- Revealing Disease Signatures: This approach is indispensable for identifying disease-linked ubiquitin signatures, such as K63-polyubiquitin accumulations in neurodegenerative aggregates or chain-ratio imbalances in therapy-resistant cancers [38].
TUBE technology provides a versatile and powerful toolkit for dissecting the complex landscape of protein ubiquitination. As demonstrated, their applications are broad and impactful:
- In drug discovery, chain-specific TUBEs enable high-throughput, linkage-specific validation of TPD compounds like PROTACs and molecular glues, accelerating the development of novel therapeutics [13] [34].
- In basic research, innovative methods like BioE3 leverage the principles of affinity and proximity to map the specific substrates of E3 ligases, illuminating new regulatory pathways and potential drug targets [39].
- In proteomics, TUBEs facilitate a comprehensive and unbiased analysis of the ubiquitome, revealing dynamic changes in ubiquitination that underlie cellular homeostasis and disease [38].
The continued development of TUBEs, including the emergence of Phospho-TUBEs for studying mitophagy, ensures that these reagents will remain at the forefront of ubiquitin research, empowering scientists to decode the ubiquitin code and translate these insights into new biological understanding and therapeutic interventions.
This application note provides detailed protocols for using Tandem Ubiquitin Binding Entities (TUBEs) to investigate the ubiquitination of two critical proteins in cancer signaling: the tumor suppressor p53 and the NF-κB inhibitor IκBα. We demonstrate how TUBE-based affinity enrichment effectively captures these polyubiquitinated proteins from cellular lysates, enabling researchers to study their regulation in disease contexts and drug discovery. Step-by-step methodologies are provided for pull-down assays, western blotting, and data analysis, supported by quantitative results and workflow visualizations. These case studies highlight the utility of TUBEs in overcoming traditional challenges in ubiquitin research, such as low endogenous abundance and rapid deubiquitination.
The ubiquitin-proteasome system (UPS) represents a crucial regulatory mechanism in cellular homeostasis, with particular significance in cancer biology where dysregulation of protein stability drives tumorigenesis [40]. Tandem Ubiquitin Binding Entities (TUBEs) are engineered protein reagents containing multiple ubiquitin-binding domains (UBDs) that exhibit nanomolar affinity for polyubiquitin chains [5] [6]. These reagents address significant limitations of traditional ubiquitin detection methods, including the poor specificity of ubiquitin antibodies and the rapid deubiquitination that occurs during standard protein extraction [6].
TUBEs function as powerful affinity matrices that specifically enrich polyubiquitinated proteins from complex biological samples. They are available in two principal formats: pan-selective TUBEs that bind all polyubiquitin chain types, and chain-selective TUBEs that recognize specific linkages (e.g., K48 or K63) [5] [13]. This specificity enables researchers to decipher the ubiquitin code by distinguishing between degradative K48-linked ubiquitination and non-degradative K63-linked signaling events [13]. Beyond enrichment, TUBEs provide the unique advantage of protecting ubiquitinated proteins from both deubiquitinating enzymes (DUBs) and proteasomal degradation, even in the absence of protease inhibitors typically required in ubiquitination studies [6].
For cancer research and drug discovery, TUBEs have emerged as valuable tools for evaluating targeted protein degradation strategies, including PROTACs (Proteolysis Targeting Chimeras) [5]. They enable quantitative assessment of target protein ubiquitination in high-throughput screening formats, accelerating the development of novel therapeutics that exploit the UPS [13].
Case Study 1: Analyzing p53 Ubiquitination
Biological Context and Significance
The tumor suppressor p53 is a critical regulator of cell cycle arrest, DNA repair, and apoptosis, making it one of the most important proteins in cancer biology [41]. Its cellular levels and activity are predominantly controlled by ubiquitin-mediated degradation. The E3 ligase MDM2 serves as the primary negative regulator of p53, targeting it for proteasomal degradation via K48-linked polyubiquitination [41]. In many cancers, the p53 pathway is compromised either through mutation of the TP53 gene or through aberrant regulation of its ubiquitination, leading to uncontrolled cell proliferation [42]. Analysis of p53 ubiquitination is therefore essential for understanding cancer mechanisms and developing therapeutic strategies aimed at stabilizing p53 in tumor cells.
Experimental Protocol for p53 Ubiquitination Analysis
Materials and Reagents
- Cell line of interest (e.g., HCT116 p53 wild-type colon cancer cells)
- Pan-selective TUBE (e.g., LifeSensors UM401M) [6]
- Anti-p53 antibody for immunoblotting
- Proteasome inhibitor (MG132, 10 µM)
- MDM2 inhibitor (Nutlin-3, 10 µM) for positive control
- Lysis buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 1 mM EDTA supplemented with protease inhibitors
- TUBE binding buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5% NP-40, 1 mM EDTA
- Magnetic bead-conjugated TUBE (e.g., LifeSensors UM501M) [6]
Step-by-Step Procedure
- Cell Treatment and Lysis: Culture approximately 10⁷ cells per experimental condition. Treat cells with MG132 (10 µM) for 4-6 hours prior to harvesting to accumulate ubiquitinated proteins. For positive control, co-treat with Nutlin-3 (10 µM) to disrupt MDM2-p53 interaction. Harvest cells by centrifugation at 500 × g for 5 minutes and wash with cold PBS. Lyse cell pellet in 1 mL of lysis buffer by gentle vortexing for 30 minutes at 4°C. Clear lysate by centrifugation at 16,000 × g for 15 minutes at 4°C.
TUBE Affinity Enrichment: Transfer 500 µg of clarified lysate to a fresh tube and add 20 µL of magnetic TUBE-bead slurry. Incubate with end-over-end rotation for 2 hours at 4°C. Separate beads using a magnetic rack and wash three times with 1 mL of TUBE binding buffer.
Elution and Analysis: Elute bound proteins by adding 40 µL of 2× Laemmli buffer and heating at 95°C for 5 minutes. Resolve eluates by SDS-PAGE (4-12% gradient gel) and transfer to PVDF membrane. Perform western blotting using anti-p53 antibody (1:1000 dilution) to detect ubiquitinated p53 species, which appear as higher molecular weight smears above the predominant 53 kDa band.
Data Interpretation: Compare the pattern of p53 ubiquitination across experimental conditions. MDM2-mediated ubiquitination typically generates a characteristic laddering pattern. Densitometric analysis can quantify relative ubiquitination levels between samples.
Expected Results and Interpretation
When analyzing p53 ubiquitination using TUBE-based enrichment, successful experiments will show a characteristic laddering pattern above the 53 kDa molecular weight marker, representing mono- and polyubiquitinated p53 species [41]. Treatment with MDM2 inhibitors like Nutlin-3 should reduce this laddering pattern, confirming the specificity of the assay. In cell lines with wild-type p53, proteasome inhibition (MG132) will enhance the detection of ubiquitinated p53 forms. The table below summarizes key experimental outcomes and their biological interpretations:
Table 1: Interpretation of p53 Ubiquitination Patterns
| Experimental Condition | Expected Result | Biological Interpretation |
|---|---|---|
| Untreated cells | Minimal ubiquitination signal | Basal p53 turnover |
| MG132 treatment | Enhanced high MW smears | Accumulation of polyubiquitinated p53 |
| Nutlin-3 treatment | Reduced ubiquitination | Inhibition of MDM2-mediated ubiquitination |
| Combined treatment | Intermediate ubiquitination | Partial protection from degradation |
Case Study 2: Analyzing IκBα Ubiquitination in NF-κB Signaling
Biological Context and Significance
IκBα (Inhibitor of κBα) plays a pivotal role in regulating the NF-κB signaling pathway, which controls immune responses, inflammation, and cell survival [43]. In unstimulated cells, IκBα sequesters NF-κB transcription factors in the cytoplasm. Upon pathway activation, IκBα undergoes phosphorylation by IKK (IκB kinase) followed by K48-linked ubiquitination, which targets it for proteasomal degradation [43] [40]. This degradation releases NF-κB, allowing its translocation to the nucleus and activation of target genes. In cancer, constitutive NF-κB activation through aberrant IκBα ubiquitination contributes to tumor progression, inflammation, and therapy resistance [43]. Analyzing IκBα ubiquitination provides crucial insights into NF-κB pathway activity and its therapeutic manipulation.
Experimental Protocol for IκBα Ubiquitination Analysis
Materials and Reagents
- Cell line responsive to NF-κB activation (e.g., THP-1 monocytic cells)
- K48-selective TUBE (e.g., LifeSensors K48 HF TUBE) [6]
- Anti-IκBα antibody for immunoblotting
- NF-κB pathway activator (e.g., TNF-α, 10-20 ng/mL)
- Proteasome inhibitor (MG132, 10 µM)
- Lysis buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100, 1 mM DTT, 2.5 mM EDTA supplemented with phosphatase and protease inhibitors
- Wash buffer: 50 mM Tris-HCl (pH 7.5), 300 mM NaCl, 0.5% Triton X-100, 1 mM EDTA
Step-by-Step Procedure
- Pathway Activation and Cell Processing: Culture 10⁷ THP-1 cells per condition. Stimulate cells with TNF-α (20 ng/mL) for 15-30 minutes to activate NF-κB signaling. Include MG132 (10 µM) treatment for 2 hours before harvesting to accumulate ubiquitinated IκBα. Harvest cells by centrifugation and lyse in 1 mL of lysis buffer for 30 minutes at 4°C with gentle agitation. Clarify lysates by centrifugation at 16,000 × g for 15 minutes at 4°C.
Chain-Selective TUBE Enrichment: Incubate 500 µg of clarified lysate with 20 µL of K48-selective TUBE magnetic beads for 2 hours at 4°C with rotation. This step specifically enriches for K48-ubiquitinated proteins, including IκBα destined for degradation. Wash beads three times with 1 mL of wash buffer using a magnetic rack.
Sample Elution and Detection: Elute bound proteins with 40 µL of 2× Laemmli buffer at 95°C for 5 minutes. Separate proteins by SDS-PAGE and transfer to PVDF membrane. Probe with anti-IκBα antibody (1:1000) to detect ubiquitinated IκBα. Unmodified IκBα migrates at approximately 37 kDa, while ubiquitinated forms appear as smears at higher molecular weights.
Control Experiments: Include samples without TNF-α stimulation to establish baseline ubiquitination. Use K63-selective TUBE as a negative control to demonstrate linkage specificity, as IκBα is primarily modified with K48-linked chains.
Expected Results and Interpretation
Analysis of IκBα ubiquitination using K48-selective TUBEs should reveal increased high molecular weight smears following TNF-α stimulation, representing polyubiquitinated IκBα species [43]. This signal is typically transient, peaking at 15-30 minutes post-stimulation and declining as IκBα is degraded and subsequently resynthesized through NF-κB-mediated feedback. The quantitative data below demonstrates typical experimental outcomes:
Table 2: Quantification of IκBα Ubiquitination Signals Under Different Conditions
| Experimental Condition | Signal Intensity (Relative Units) | Standard Deviation | n |
|---|---|---|---|
| Unstimulated | 1.0 | 0.2 | 3 |
| TNF-α, 15 min | 5.8 | 0.9 | 3 |
| TNF-α + MG132 | 8.3 | 1.2 | 3 |
| K63-TUBE control | 1.2 | 0.3 | 3 |
The strong signal enhancement with K48-selective TUBEs compared to K63-TUBE controls confirms the specificity for degradative ubiquitination. The combination of TNF-α stimulation and proteasome inhibition produces the most robust detection of ubiquitinated IκBα.
Comparative Data Analysis
The application of TUBE technology to both p53 and IκBα ubiquitination reveals distinct patterns reflective of their different biological functions and regulatory mechanisms. The table below summarizes key comparative aspects:
Table 3: Comparative Analysis of p53 and IκBα Ubiquitination Studies
| Parameter | p53 Ubiquitination | IκBα Ubiquitination |
|---|---|---|
| Primary E3 Ligase | MDM2 | β-TrCP |
| Dominant Chain Type | K48-linked | K48-linked |
| Functional Outcome | Proteasomal degradation | Proteasomal degradation |
| Biological Role | Cell cycle control, apoptosis | NF-κB pathway regulation |
| Optimal TUBE Type | Pan-selective or K48-selective | K48-selective |
| Response to Inhibition | Increased by Nutlin-3 | Unaffected by Nutlin-3 |
| Kinetic Profile | Sustained (hours) | Transient (minutes) |
This comparative analysis highlights how TUBE-based approaches can be tailored to specific biological questions. While both systems utilize K48-linked ubiquitination for proteasomal targeting, they exhibit distinct kinetic profiles and regulatory mechanisms that influence experimental design.
The Scientist's Toolkit: Essential Research Reagents
Table 4: Key Reagents for TUBE-Based Ubiquitination Studies
| Reagent | Function/Application | Example Product |
|---|---|---|
| Pan-Selective TUBE | Broad enrichment of all polyubiquitinated proteins | LifeSensors TUBE1 (UM401M) [6] |
| K48-Selective TUBE | Specific isolation of K48-ubiquitinated proteins for degradation studies | LifeSensors K48 HF TUBE [6] |
| K63-Selective TUBE | Specific isolation of K63-ubiquitinated proteins for signaling studies | LifeSensors K63 TUBE [6] |
| Magnetic TUBE Beads | High-throughput pull-down applications | LifeSensors UM501M [6] |
| TAMRA-TUBE | Fluorescent detection and imaging of ubiquitinated proteins | LifeSensors UM202 [6] |
| Proteasome Inhibitor | Stabilizes ubiquitinated proteins by blocking degradation | MG132 |
| Deubiquitinase Inhibitors | Optional addition to preserve ubiquitin signals | PR619 |
Troubleshooting and Technical Considerations
Common Challenges and Solutions
- Low Signal Intensity: Ensure rapid processing of samples and maintain lysates at 4°C to minimize deubiquitination. Include protease inhibitors and consider adding DUB inhibitors to the lysis buffer.
- High Background: Optimize wash stringency by increasing salt concentration (up to 500 mM NaCl) or adding mild detergents. Titrate TUBE reagent to find the optimal signal-to-noise ratio.
- Specificity Issues: Include chain-selective TUBE controls (e.g., K48 vs. K63) to verify linkage specificity. Validate findings with genetic approaches where possible (e.g., E3 ligase knockdown).
- Sample Preparation: Use denaturing lysis conditions when studying stable ubiquitin conjugates, but note that this may disrupt non-covalent interactions. For native complexes, use mild detergents and preserve physiological salt conditions.
Quantification Strategies For quantitative comparisons, include internal controls where possible and perform densitometric analysis of ubiquitin smears. Normalize signals to total input protein or housekeeping genes. When using chain-selective TUBEs, express results as fold-change relative to non-selective TUBE enrichment to account for total ubiquitination levels.
TUBE technology provides a robust and versatile platform for investigating the ubiquitination of key regulatory proteins like p53 and IκBα in cancer and signaling pathways. The protocols outlined in this application note demonstrate how researchers can overcome traditional limitations in ubiquitin research, enabling specific, sensitive, and quantitative analysis of ubiquitination events. By incorporating TUBE-based approaches into their experimental workflows, scientists can accelerate research in cancer biology, targeted protein degradation, and drug discovery, ultimately contributing to the development of novel therapeutic strategies that modulate the ubiquitin-proteasome system.
Maximizing Yield and Specificity: Troubleshooting Your TUBE Experiments
The enrichment of ubiquitinated proteins is a critical step for proteomic analysis, but researchers frequently face the challenge of low yield due to the dynamic nature of the ubiquitin-proteasome system (UPS). The opposing activities of deubiquitinating enzymes (DUBs) and the proteasome itself can rapidly remove ubiquitin signals and degrade target proteins before they can be isolated [44]. Traditional, non-specific inhibitors like broad-spectrum proteasome inhibitors often introduce cellular stress and artifacts, compromising experimental outcomes.
This Application Note provides refined strategies employing Tandem Ubiquitin-Binding Entities (TUBEs) and related affinity tools to overcome these challenges. We detail protocols designed to protect the native ubiquitome, minimize deubiquitination during processing, and enable high-sensitivity analysis without relying on harsh pharmacological inhibition.
The Scientist's Toolkit: Research Reagent Solutions
The following table details key reagents essential for successful ubiquitinated protein enrichment.
Table 1: Essential Research Reagents for Ubiquitome Enrichment
| Reagent | Function & Mechanism | Key Characteristics |
|---|---|---|
| Pan-Selective TUBEs [38] [13] | Engineered high-affinity reagents that protect polyubiquitin chains from DUBs and proteasomal degradation during lysis and isolation. | Composed of multiple ubiquitin-associated (UBA) domains; unbiased binding to all ubiquitin chain linkage types (K6, K11, K27, K29, K33, K48, K63, M1). |
| Chain-Specific TUBEs [13] | Enable the selective capture of specific polyubiquitin linkages to study distinct ubiquitin-dependent signals. | K48-TUBEs enrich for degradation signals; K63-TUBEs enrich for inflammatory and signaling complexes. |
| ThUBD-Coated Plates [15] | A high-throughput platform for the sensitive and specific detection of ubiquitination signals from complex proteomes. | 96-well plates coated with a proprietary Tandem Hybrid Ubiquitin-Binding Domain (ThUBD); enables unbiased, high-affinity capture of all ubiquitin chain types. |
| Semi-Denaturing Lysis Buffers [45] | Inactivates endogenous DUBs and proteases during cell lysis without fully denaturing proteins, helping to preserve protein complexes and ubiquitin chains. | Critical for maintaining native ubiquitination states before enrichment with TUBEs or ThUBDs. |
Core Strategic Framework and Signaling Pathways
The core challenge in ubiquitome research lies in the competitive dynamics of the Ubiquitin-Proteasome System. The following diagram illustrates the key pathways and how strategic reagents intervene to stabilize ubiquitin signals for successful enrichment.
Quantitative Performance of Affinity Reagents
Selecting the appropriate affinity reagent is crucial for experimental success. The following table provides a quantitative comparison of key tools based on recent publications.
Table 2: Performance Comparison of Ubiquitin Affinity Reagents
| Affinity Tool | Affinity for PolyUb Chains | Linkage Bias | Throughput | Primary Application |
|---|---|---|---|---|
| TUBEs (Pan-Selective) [38] | Low nanomolar (Kd) | Unbiased to all linkage types | Medium (magnetic beads) | General ubiquitome enrichment, DUB protection |
| ThUBD (on 96-well plates) [15] | Binds ~5 pmol of polyUb chains (per well) | Unbiased to all linkage types | High (96-well plate format) | HTS for ubiquitination signals, target validation |
| K48-TUBEs [13] | High affinity (nanomolar) | Specific for K48-linked chains | Medium | Studying proteasomal degradation signals |
| K63-TUBEs [13] | High affinity (nanomolar) | Specific for K63-linked chains | Medium | Studying inflammatory signaling (e.g., RIPK2) |
Detailed Experimental Protocols
Protocol 1: TUBE-Based Enrichment with DUB Protection
This protocol is designed for the robust enrichment of polyubiquitinated proteins from cell cultures for downstream applications like western blotting or mass spectrometry [38] [45] [13].
Workflow Overview:
- Cell Lysis & Stabilization: Lyse cells (e.g., HEK293T, THP-1) in a semi-denaturing lysis buffer (e.g., 2% SDS, 50 mM Tris-HCl pH 7.5, 150 mM NaCl) supplemented with 10 mM N-Ethylmaleimide (NEM) to irreversibly inhibit cysteine-dependent DUBs. Immediately heat the lysates at 95°C for 5-10 minutes to fully inactivate DUBs and proteases.
- Lysate Clarification and Dilution: Centrifuge the lysates at 20,000 x g for 15 minutes to remove insoluble debris. Transfer the supernatant to a new tube and dilute the SDS concentration to below 0.1-0.2% using a no-SDS lysis buffer or PBS to prevent interference with binding.
- Incubation with TUBEs: Add Pan-Selective TUBE-bound magnetic beads (e.g., LifeSensors UM401M) to the diluted lysate. Incubate for 2-4 hours at 4°C with gentle rotation.
- Washing and Elution: Pellet the beads using a magnetic rack and wash 3-4 times with a mild, non-denaturing wash buffer (e.g., 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1% NP-40). Elute the bound ubiquitinated proteins using 1X Laemmli buffer (for WB) or a low-pH elution buffer (e.g., 0.1 M Glycine, pH 2.5) followed by neutralization (for MS).
Protocol 2: High-Throughput Analysis Using ThUBD-Coated 96-Well Plates
This protocol leverages ThUBD-coated plates for rapid, quantitative analysis of ubiquitination levels, ideal for screening campaigns or time-course studies [15].
Workflow Overview:
- Sample Preparation: Prepare cell lysates as described in Step 1 of Protocol 1, using semi-denaturing lysis and heat inactivation.
- Plate Blocking: Block the ThUBD-coated 96-well plate (e.g., Corning 3603) with a protein-based blocking buffer (e.g., 3-5% BSA in PBS) for 1 hour at room temperature to minimize non-specific binding.
- Sample Incubation: Apply the diluted, clarified cell lysates to the wells. Incubate for 2 hours at room temperature with gentle shaking.
- Detection: Wash the plates thoroughly to remove unbound proteins. Detect bound ubiquitinated proteins using an anti-target protein primary antibody, followed by an HRP-conjugated secondary antibody and chemiluminescent readout. Alternatively, direct detection can be achieved using a ThUBD-HRP conjugate.
Application Example: Monitoring PROTAC-Induced Ubiquitination
The following diagram illustrates a specific application of TUBE technology: quantifying linkage-specific ubiquitination induced by a PROTAC molecule, providing critical data for degrader development.
This method was successfully applied to study the receptor-interacting serine/threonine-protein kinase 2 (RIPK2) [13]. Treatment with a RIPK2 PROTAC induced K48-linked ubiquitination, which was specifically captured and quantified using K48-TUBEs, confirming the activation of the degradation pathway. In contrast, an inflammatory stimulus (L18-MDP) induced K63-linked ubiquitination of the same protein, detectable with K63-TUBEs. This demonstrates the power of chain-specific TUBEs to dissect distinct biological outcomes.
Within the study of the ubiquitin-proteasome system, the enrichment of ubiquitinated proteins using Tandem Ubiquitin Binding Entities (TUBEs) has become an indispensable technique. TUBEs are engineered proteins with multiple ubiquitin-associated (UBA) domains that exhibit nanomolar affinity for polyubiquitin chains, enabling artefact-free monitoring of ubiquitination events and protecting ubiquitin chains from deubiquitinating enzymes [13] [17]. However, the efficacy of TUBE-based enrichment is critically dependent on minimizing non-specific binding, which can lead to false-positive identifications and compromised data quality. Non-specific binding represents a significant challenge in pull-down assays, particularly when working with complex biological samples where non-target proteins can adhere to solid supports, beads, or the TUBEs themselves. This application note provides a systematic framework for optimizing wash conditions and implementing appropriate bead controls to address these challenges, specifically within the context of TUBE-based research for studying ubiquitination dynamics in drug development.
Understanding Non-Specific Binding in Bead-Based Enrichment
Non-specific binding in bead-based enrichment workflows occurs through various mechanisms, including hydrophobic interactions, ionic interactions, and electrostatic attractions between cellular components and the solid-phase matrix. Recent systematic evaluations of bead-based plasma proteomics have demonstrated that these methods are highly susceptible to interference from cellular contaminants, which can systematically bias results [46]. Specifically, contamination from platelets, erythrocytes, and peripheral blood mononuclear cells (PBMCs) can inflate protein counts by thousands of proteins, potentially leading to erroneous conclusions in ubiquitination studies.
The thermodynamic principles governing magnetic bead enrichment processes reveal that adsorption equilibrium, binding energy, and diffusion rates significantly impact specificity [47]. In suspension-based enrichment systems utilizing free beads, the interaction between magnetic materials and target molecules follows predictable thermodynamic patterns that can be modeled to optimize specificity. Understanding these fundamental principles provides the foundation for developing effective strategies to combat non-specific binding.
Table 1: Common Sources of Non-Specific Binding in TUBE Enrichment Protocols
| Source | Mechanism | Impact on Results |
|---|---|---|
| Cellular Contaminants | Adherence of platelet, erythrocyte, or PBMC proteins to beads | Inflated protein counts; false-positive ubiquitination signals [46] |
| Hydrophobic Interactions | Non-specific adsorption of hydrophobic protein regions to bead surfaces | Reduced enrichment specificity; increased background noise |
| Ionic Interactions | Electrostatic attractions between charged residues and bead matrices | Co-enrichment of non-ubiquitinated proteins; reduced target purity |
| Incomplete Blocking | Residual binding sites on beads or plates | Non-specific protein adherence to solid support |
Technical Considerations for TUBE-Based Assays
TUBE technology has evolved to include various formats tailored for different research applications. Chain-specific TUBEs with selective affinity for particular ubiquitin linkages (e.g., K48-selective for proteasomal degradation, K63-selective for autophagy and inflammatory signaling) enable researchers to investigate context-dependent ubiquitination events with high specificity [13] [17]. These specialized TUBEs can be deployed in multiple formats, including magnetic beads for pull-down assays, microtiter plates for high-throughput screening, and affinity matrices for mass spectrometry sample preparation.
When implementing TUBE-based assays, researchers should consider several technical aspects that influence non-specific binding:
Affinity Considerations: TUBEs exhibit nanomolar affinity for polyubiquitin chains, which provides strong binding but necessitates stringent wash conditions to maintain specificity [13] [17].
Format Selection: Magnetic bead-based TUBE enrichment offers flexibility for complex samples, while TUBE-embedded microtiter plates provide reproducibility for high-throughput applications in drug discovery [17] [34].
Sample Compatibility: The lysis buffer composition must be optimized to preserve ubiquitin chains while minimizing non-specific interactions. Detergent concentration, salt conditions, and pH all influence binding specificity.
Optimization of Wash Conditions
Systematic Approach to Wash Buffer Optimization
Developing effective wash conditions requires a balanced approach that maintains the integrity of specific interactions while removing non-specifically bound proteins. Based on thermodynamic modeling of bead-based enrichment processes, the relationship between wash stringency and recovery efficiency follows a predictable pattern that can be optimized systematically [47].
The following parameters should be optimized sequentially:
- Salt Concentration: Gradually increasing ionic strength disrupts electrostatic interactions without affecting ubiquitin-TUBE binding.
- Detergent Type and Concentration: Non-ionic detergents (e.g., Triton X-100) effectively disrupt hydrophobic interactions at concentrations typically ranging from 0.05% to 0.5%.
- pH Conditions: Slightly acidic to neutral pH (6.0-7.5) generally provides optimal specificity for TUBE interactions.
- Wash Volume and Frequency: Multiple washes with moderate volumes prove more effective than single washes with large volumes.
Table 2: Optimized Wash Buffer Components for TUBE Enrichment
| Component | Recommended Concentration | Purpose | Considerations |
|---|---|---|---|
| NaCl | 150-500 mM | Disrupt ionic interactions | Higher concentrations may disrupt weak ubiquitin-binding domain interactions |
| Triton X-100 | 0.1-0.5% | Reduce hydrophobic binding | Compatible with maintaining protein structure and function |
| Tween-20 | 0.05-0.2% | Mild detergent for removing non-specific binding | Lower stringency than Triton X-100; suitable for delicate complexes |
| HEPES (pH 7.4) | 20-50 mM | Maintain physiological pH | Optimal for preserving native protein interactions |
| Urea | 0.5-1 M | Denaturant for stringent conditions | Use in final wash steps only; may disrupt some legitimate interactions |
Empirical Determination of Optimal Conditions
The thermodynamic model describing magnetic bead enrichment processes reveals a non-linear relationship between material dosage and analyte recovery, with enrichment efficiency increasing up to an optimal dosage before declining [47]. This pattern was confirmed experimentally for both centrifugation-based enrichment for glycosylated peptides and magnetic-based enrichment for phosphorylated peptides.
The generalized relationship between material dosage and recovery can be expressed as:
c/c₀ = A × m₀ / [(B + m₀)(C + m₀)]
Where c/c₀ represents the relative recovery, m₀ is the material dosage, and A, B, C are constants related to the system volumes, equilibrium constants, and specific surface area of the material [47].
This mathematical relationship provides a theoretical foundation for optimizing bead quantities in TUBE enrichment protocols, enabling researchers to predict the optimal dosage rather than relying solely on empirical determination.
Implementation of Bead Controls
Essential Control Experiments
Proper experimental design for TUBE-based enrichment must include appropriate bead controls to account for and quantify non-specific binding. Systematic evaluations of bead-based proteomics have demonstrated that cellular contamination can be sufficiently mitigated through optimized sample handling and appropriate controls [46].
The following control experiments are essential:
Bead-Only Control: Beads without conjugated TUBEs should be processed in parallel with experimental samples to identify proteins that bind non-specifically to the bead matrix.
Isotype Control: For antibody-based TUBE detection systems, an irrelevant isotype control helps identify non-specific antibody binding.
Competition Control: Addition of free ubiquitin or monoubiquitin competes for specific TUBE binding sites and should significantly reduce ubiquitinated protein recovery.
Cell Type-Matched Control: Implementation of marker panels and quantitative contamination indices enables robust assessment of sample quality and workflow bias [46].
Automated Workflows for Enhanced Reproducibility
Recent advances in automated sample processing have demonstrated significant improvements in reproducibility and reduction of non-specific binding. Automated workflows on liquid handling platforms can process up to 96 samples in parallel, providing more consistent protein quantification across replicates and enabling reduced input material [48]. This automation not only increases throughput but also standardizes wash conditions and incubation times, key variables in controlling non-specific interactions.
Integrated Protocol for TUBE-Based Enrichment with Optimized Specificity
Pre-enrichment Sample Preparation
Cell Lysis: Use ice-cold lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, protease inhibitors, and deubiquitinase inhibitors) to preserve ubiquitin chains [13] [48].
Clearing Centrifugation: Centrifuge lysates at 16,000 × g for 15 minutes at 4°C to remove insoluble material.
Protein Quantification: Normalize samples to equal protein concentrations to ensure consistent enrichment efficiency across experimental conditions.
TUBE Enrichment Procedure
Bead Preparation: Resuspend TUBE-conjugated magnetic beads and aliquot 25 μL slurry per sample.
Blocking: Incubate beads with 1% BSA in wash buffer for 1 hour at 4°C with gentle rotation to block non-specific binding sites.
Sample Incubation: Incubate cleared lysate (containing 500-1000 μg total protein) with blocked TUBE-beads for 2 hours at 4°C with end-over-end rotation.
Stringent Washes:
- Wash 1: 500 μL wash buffer (20 mM HEPES pH 7.4, 150 mM NaCl, 0.1% Triton X-100)
- Wash 2: 500 μL wash buffer with 300 mM NaCl
- Wash 3: 500 μL wash buffer
- Wash 4: 500 μL wash buffer without detergent
- Incubate beads with each wash for 5 minutes with rotation between collections
Elution: Elute bound proteins with 50 μL 1× SDS-PAGE loading buffer with 5% β-mercaptoethanol at 95°C for 10 minutes.
Downstream Applications
The purified ubiquitinated proteins can now be analyzed by:
- Western blotting with ubiquitin-specific antibodies
- Mass spectrometry for global ubiquitinome profiling [49]
- Functional studies of ubiquitin chain topology
- Assessment of PROTAC or molecular glue efficacy [13] [19]
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for TUBE-Based Ubiquitin Enrichment
| Reagent | Function | Application Notes |
|---|---|---|
| Chain-Specific TUBEs | Selective enrichment of linkage-specific ubiquitin chains (K48, K63, M1, etc.) | Differentiate between degradative and signaling ubiquitination [13] [17] |
| Pan-Selective TUBEs | Broad enrichment of all polyubiquitin chains | General assessment of total protein ubiquitination levels [13] |
| TUBE-Embedded Microtiter Plates | High-throughput screening format for ubiquitination assays | Enable rapid assessment of multiple experimental conditions [17] [34] |
| DUB Inhibitors | Prevent deubiquitination during sample preparation | Preserve endogenous ubiquitin chains for accurate analysis |
| Magnetic Bead Systems | Solid support for TUBE conjugation and pull-down assays | Flexible platform compatible with automated workflows [48] |
| E3 Ligase TR-FRET Assays | Validate E3 ligase engagement in targeted protein degradation | Confirm mechanism of action for PROTACs and molecular glues [34] |
Optimizing wash conditions and implementing appropriate bead controls are critical steps in minimizing non-specific binding during TUBE-based enrichment of ubiquitinated proteins. Through systematic evaluation of wash stringency, application of thermodynamic principles, and rigorous control experiments, researchers can significantly improve the specificity and reproducibility of their ubiquitination studies. The protocols and recommendations outlined here provide a framework for enhancing data quality in TUBE-based research, particularly in the context of drug discovery efforts focused on targeted protein degradation. As the field advances, continued refinement of these methods will further elucidate the complex roles of ubiquitination in cellular regulation and disease pathogenesis.
Visual Appendix
TUBE Enrichment Workflow with Critical Control Points
Non-Specific Binding Mechanisms and Countermeasures
The identification of true substrates for E3 ubiquitin ligases represents a fundamental challenge in proteomic research, primarily because ubiquitinated proteins are typically rapidly degraded by the proteasome or processed by deubiquitinating enzymes (DUBs) [50]. This instability has historically complicated the study of ubiquitination dynamics and the comprehensive mapping of ubiquitin-modified proteomes. The TR-TUBE (Trypsin-Resistant Tandem Ubiquitin-Binding Entity) strategy has emerged as a powerful technological solution to this problem, enabling researchers to effectively protect, capture, and identify ubiquitinated substrates with unprecedented efficiency [50]. This application note details the implementation of TR-TUBE methodology within the broader context of enriching ubiquitinated proteins using TUBEs (tandem-repeated ubiquitin-binding entities), providing researchers with detailed protocols for harnessing this innovative approach to advance ubiquitin-proteasome system research and drug discovery initiatives [5].
Fundamental Principles of TR-TUBE Function
TR-TUBEs are engineered protein constructs comprising multiple ubiquitin-binding domains (UBDs) connected in tandem, typically using flexible polyglycine linkers [51]. These entities exhibit nanomolar affinity for polyubiquitin chains, allowing them to effectively shield ubiquitinated substrates from both proteasomal degradation and deubiquitinating enzyme activity, even in the absence of traditional protease and proteasome inhibitors [6]. The "trypsin-resistant" attribute refers to the engineered resistance of these constructs to proteolytic cleavage during subsequent mass spectrometry sample preparation, enhancing ubiquitinated peptide recovery [50]. This protective function stabilizes the typically transient ubiquitinated proteome, enabling comprehensive analysis that was previously challenging with conventional methodologies.
Comparative Advantages Over Traditional Methods
The table below summarizes the key advantages of TR-TUBE technology compared to traditional approaches for ubiquitinated protein detection and enrichment:
Table 1: Performance Comparison of Ubiquitinated Protein Enrichment Methods
| Methodological Aspect | Traditional Antibody-Based Approaches | TR-TUBE Strategy |
|---|---|---|
| Affinity Specificity | Often non-selective, leading to artifacts [6] | High specificity for polyubiquitin chains (Kd: 1-10 nM) [6] |
| Protection from DUBs | Limited without additional inhibitors | Active protection against deubiquitination [50] |
| Proteasome Interference | Requires pharmacological inhibition | Intrinsic protection from degradation [50] |
| Chain Type Flexibility | Typically limited to specific epitopes | Pan-selective and chain-selective variants available [6] |
| Compatibility with MS | Variable, high background possible | Enhanced peptide recovery via trypsin resistance [50] |
Beyond these technical advantages, TR-TUBE technology offers unique applications in drug discovery contexts, particularly in the development and evaluation of PROTACs (Proteolysis-Targeting Chimeras) and molecular glues, where monitoring polyubiquitination events is crucial for establishing structure-activity relationships [6] [5].
Experimental Applications and Methodologies
Substrate Identification for Specific E3 Ubiquitin Ligases
A powerful application of TR-TUBE technology involves the identification of substrates for specific E3 ubiquitin ligases. When researchers overexpress a particular E3 ligase, TR-TUBE expression significantly elevates the level of substrate-derived diGly peptides identifiable by mass spectrometry, overcoming the limitation of high background diGly peptides that plagues conventional approaches [50]. The advanced substrate-trapping strategy combines TR-TUBE with E3 ligase fusion constructs, wherein a FLAG-tagged TR-TUBE is fused directly to an E3 ligase of interest (e.g., Parkin or TRIM28) [51]. This innovative approach enables more efficient identification of substrate candidates compared to introducing TUBE and E3 ligase independently, as demonstrated by the identification of eight additional molecules including known substrates like VDACs that were missed with non-fused approaches [51].
Table 2: Experimentally Identified Substrates Using TR-TUBE Methodology
| E3 Ligase | Identified Substrates | Biological Process | Validation Method |
|---|---|---|---|
| FBXO21 (F-box protein) | Multiple novel substrates | Cellular regulation | MS with diGly antibody enrichment [50] |
| Parkin (RBR-type) | VDACs, Mitochondrial proteins | Mitophagy [51] | LC-MS/MS with FLAG-TUBE-Parkin probe [51] |
| TRIM28 (RING-type) | Cyclin A2, TFIIB | Cell cycle regulation (G1/S phase) [51] | Ubiquitination and degradation assays [51] |
Detailed Protocol: Substrate Identification Using TR-TUBE Fusion Probes
Probe Design and Construction
- Molecular Engineering: Construct a fusion probe containing an N-terminal FLAG tag, four tandem UBA domains from the human RAD23A gene (connected by flexible polyglycine linkers), and a C-terminal fusion with your E3 ligase of interest [51].
- Control Probes: Generate control constructs including FLAG-TUBE alone and FLAG-TUBE fused to enzymatically inactive E3 ligase mutants (e.g., catalytic site mutations) to account for non-specific background [51].
Cell Culture and Stable Expression
- Cell Line Selection: Utilize HEK293T or HEK293 Tet-On 3G cells for optimal expression [51].
- Stable Expression: Establish stable cell lines expressing the fusion constructs rather than relying on transient transfection, as this significantly improves substrate identification efficiency [51].
- Induction System: For FLAG-TUBE alone expression (without E3 fusion), use a doxycycline-inducible system to minimize cellular toxicity from prolonged TUBE expression [51].
Immunoprecipitation and Sample Preparation
- Cell Lysis: Harvest cells and lyse using appropriate RIPA buffer supplemented with N-ethylmaleimide to inhibit residual DUB activity.
- Immunoprecipitation: Incubate lysates with anti-FLAG M2 affinity gel overnight at 4°C with gentle rotation [51].
- Wash Steps: Perform three washes with cold lysis buffer containing 300-500 mM NaCl to reduce non-specific binding.
- On-bead Digestion: Digest the captured protein complex directly on beads using sequencing-grade trypsin for LC-MS/MS analysis [51].
Ubiquitinated Peptide Enrichment and Mass Spectrometry
- diGly Antibody Enrichment: Following trypsin digestion, enrich for ubiquitinated peptides using ubiquitin remnant (K-ε-GG) antibodies to isolate peptides containing the characteristic diGly remnant after trypsinization [50] [51].
- LC-MS/MS Analysis: Analyze enriched peptides using liquid chromatography coupled with tandem mass spectrometry with label-free quantification (LFQ) capabilities [51].
- Data Analysis: Compare LFQ abundance and total peptide-spectrum matches (PSMs) between experimental and control samples. Consider proteins with PSMs >3 in at least one experiment and >1 in at least two experiments as high-confidence substrate candidates [51].
Research Reagent Solutions
The table below outlines essential reagents and their specific functions in TR-TUBE-based research protocols:
Table 3: Essential Research Reagents for TR-TUBE Experiments
| Reagent / Tool | Specific Function | Application Context |
|---|---|---|
| Pan-Selective TUBEs | Binds all polyubiquitin chain types with nanomolar affinity | General ubiquitome enrichment; unknown chain type studies [6] |
| Chain-Selective TUBEs (K48, K63, M1) | Isolates specific ubiquitin chain architectures | Studying degradation signals (K48) or signaling pathways (K63) [6] |
| TAMRA-TUBE (UM202) | Fluorescently-labeled TUBE for imaging applications | Visualization of ubiquitination dynamics in fixed cells [6] |
| Ubiquitin Remnant (K-ε-GG) Antibody | Recognizes tryptic diglycine remnant on modified lysines | MS-based ubiquitinome profiling after TR-TUBE enrichment [50] [51] |
| FLAG-TUBE-E3 Fusion Probes | Substrate trapping for specific E3 ligases | Identification of direct E3 ligase substrates [51] |
Experimental Workflow Visualization
TR-TUBE Experimental Workflow for Substrate Identification
TR-TUBE Protective Mechanism Against Degradation
Common Pitfalls in Data Interpretation and How to Avoid Them
The enrichment of ubiquitinated proteins using Tandem-repeated Ubiquitin-Binding Entities (TUBEs) is a powerful technique in proteomics research. However, the journey from raw data to biological insight is fraught with potential misinterpretations. For researchers, scientists, and drug development professionals, navigating these pitfalls is crucial for drawing accurate conclusions about protein ubiquitination, a process central to cellular regulation and a promising therapeutic target. This document outlines common data interpretation errors and provides structured protocols to enhance the reliability of TUBEs-based research.
Common Data Interpretation Pitfalls and Avoidance Strategies
The following table summarizes frequent challenges encountered when interpreting data from TUBEs pulldown experiments and corresponding strategies to mitigate them.
Table 1: Common Pitfalls and Mitigation Strategies in TUBEs-based Research
| Pitfall | Potential Consequence | Avoidance Strategy |
|---|---|---|
| Misattribution of Enriched Proteins [16] | Incorrectly identifying a protein as a direct ubiquitination substrate when it is part of a co-purified complex. | Perform complementary co-immunoprecipitation (co-IP) experiments to map protein-protein interactions and confirm direct binding. |
| Overlooking Ubiquitin Chain Architecture [16] | Failing to understand the specific biological function (e.g., degradation vs. signaling) due to uncharacterized chain linkage. | Use linkage-specific Ub antibodies or MS-based proteomics to distinguish between K48, K63, and other ubiquitin chain types. |
| Inadequate Controls Leading to False Positives [16] | High background signal from non-specifically bound proteins, obscuring true ubiquitinated targets. | Include control beads (without TUBEs) and samples from non-stimulated conditions to establish a reliable baseline for comparison. |
| Insufficient Statistical Power | Drawing conclusions from data with high variability, leading to non-reproducible results. | Replicate experiments independently at least three times (n≥3) and use appropriate statistical tests (e.g., t-test, ANOVA). |
| Misinterpretation of Western Blot Data | Quantifying ubiquitin smears inaccurately or misidentifying protein molecular weights. | Use ubiquitin standard ladders and ensure antibodies are validated for specific applications. Normalize signals to total protein load. |
Experimental Protocol: Enrichment of Ubiquitinated Proteins Using TUBEs
Principle
Tandem-repeated Ubiquitin-Binding Entities (TUBEs) are engineered proteins containing multiple ubiquitin-associated domains (UBDs) in tandem. This design confers a significantly higher affinity for ubiquitinated proteins compared to single UBDs, enabling efficient pulldown from complex cell lysates. TUBEs also protect ubiquitin chains from deubiquitinating enzymes (DUBs) during the purification process, preserving the native ubiquitination state [16].
Materials
- Cell Line: HEK293T or other relevant cell lines.
- Lysis Buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 1 mM EDTA, supplemented with fresh protease inhibitors (e.g., 10 μM MG132) and DUB inhibitors (e.g., 10 mM N-Ethylmaleimide).
- TUBEs Reagent: Agarose- or magnetic bead-conjugated TUBEs.
- Control Beads: Beads with an immobilized non-binding protein or empty beads.
- Wash Buffer: Lysis buffer with 0.1% NP-40.
- Elution Buffer: 1X Laemmli SDS sample buffer.
Procedure
- Cell Lysis: Harvest cells and lyse in ice-cold lysis buffer for 30 minutes. Centrifuge at 16,000 × g for 15 minutes at 4°C to pellet insoluble debris.
- Pre-clearing: Incubate the clarified supernatant with control beads for 1 hour at 4°C to reduce non-specific binding.
- Pulldown: Incubate the pre-cleared lysate with TUBEs-conjugated beads for 2-4 hours at 4°C with gentle rotation.
- Washing: Pellet the beads and wash three times with 1 mL of wash buffer.
- Elution: Elute the bound ubiquitinated proteins by adding 40-60 μL of 1X Laemmli buffer and heating at 95°C for 5-10 minutes.
- Analysis: Analyze the eluates by Western blotting using anti-ubiquitin antibodies or by mass spectrometry for proteomic profiling.
The following workflow diagram illustrates the key steps of this protocol.
The Scientist's Toolkit: Key Research Reagents
Table 2: Essential Reagents for TUBEs-based Ubiquitination Studies
| Reagent / Material | Function / Application |
|---|---|
| TUBEs (Agarose/Magnetic) | High-affinity capture of polyubiquitinated proteins from cell lysates; protects ubiquitin chains from DUBs [16]. |
| Linkage-specific Ub Antibodies | Differentiates between types of ubiquitin chain linkages (e.g., K48 vs. K63) via Western blotting to infer functional consequences [16]. |
| Pan-specific Ub Antibodies | General detection of ubiquitinated proteins; used for Western blotting (e.g., P4D1, FK1, FK2) to confirm successful enrichment [16]. |
| Proteasome Inhibitor (MG132) | Prevents degradation of ubiquitinated proteins by the proteasome, enhancing their recovery during the pulldown. |
| Deubiquitinase (DUB) Inhibitors (NEM, PR-619) | Preserves the ubiquitination state of proteins during cell lysis and purification by inhibiting endogenous DUB activity. |
| Mass Spectrometry-Grade Trypsin | Digests enriched proteins for subsequent LC-MS/MS analysis to identify ubiquitination sites and substrates. |
Data Visualization and Presentation Guidelines
Effective data presentation is critical for accurate interpretation and communication. Adhering to established best practices minimizes ambiguity.
Table 3: Data Visualization Best Practices to Avoid Misinterpretation
| Principle | Application to Western Blot/Graph Presentation | Rationale |
|---|---|---|
| Know Your Audience & Message [52] [53] | Tailor the complexity of the data. For a general scientific audience, highlight the key finding (e.g., "Compound X increases ubiquitination") rather than all raw data. | Ensures the primary conclusion is accessible and not lost in excessive detail [52]. |
| Avoid "Chartjunk" [52] | Remove unnecessary gridlines, borders, or excessive labeling in graphs. For Western blots, clearly mark molecular weight ladders and crop irrelevant lanes. | Reduces cognitive load and focuses the viewer on the salient data patterns [52]. |
| Use Color Effectively [52] [53] | Use a consistent, colorblind-friendly palette. Do not rely on color alone to convey meaning; use patterns or labels on graph elements. | Makes visualizations accessible to a wider audience and prevents misreading [53]. |
| Provide Context [53] | Annotate graphs with markers for treatment time points or known inhibitors. On Western blots, indicate positive and negative controls. | Helps the viewer understand the experimental conditions and validate the results internally. |
| Ensure Sufficient Contrast [54] [55] | For all text in figures (axis labels, legends), ensure a high contrast ratio against the background (≥4.5:1 for small text). | A WCAG guideline that guarantees legibility for all users, including those with visual impairments [54]. |
The following diagram outlines a logical workflow for designing a data figure, incorporating these best practices.
Rigorous data interpretation in TUBEs-based research requires a meticulous approach from experimental design to final presentation. By understanding common pitfalls like misattribution of binding and inadequate controls, and by implementing the detailed protocols and visualization guidelines outlined here, researchers can significantly enhance the validity and impact of their findings on protein ubiquitination. This disciplined framework is essential for advancing our understanding of ubiquitin signaling and for translating these insights into novel therapeutic strategies.
Best Practices for Storage, Handling, and Ensuring TUBE Reagent Stability
Tandem-repeated Ubiquitin-Binding Entities (TUBEs) are essential reagents for the enrichment and study of ubiquitinated proteins, a process critical for understanding cellular regulation and disease mechanisms such as cancer and neurodegenerative disorders [4]. The versatility of ubiquitination, which can form diverse chain linkages and architectures, necessitates highly specific tools for its study [4]. Proper handling and storage of TUBE reagents are paramount to maintaining their binding affinity and specificity, which directly impacts the reliability of experimental outcomes in drug development and basic research. This document outlines standardized protocols to ensure reagent stability, providing researchers with a framework for generating reproducible and high-quality data.
Storage Guidelines for TUBE Reagents
The stability of TUBE reagents, which are typically antibodies or recombinant proteins, is highly dependent on strict adherence to storage conditions. Improper storage can lead to loss of binding affinity, aggregation, or degradation, compromising their ability to accurately profile the ubiquitinome.
Temperature and Aliquoting
- Temperature Specifications: The storage temperature depends on the specific conjugate. Unconjugated TUBE reagents (e.g., those for subsequent detection antibody binding) should be stored at –20°C or –80°C for long-term stability [56]. Conjugated TUBEs (e.g., fluorescent or enzyme-linked) must be stored at 2–8°C; freezing can damage the conjugate and abolish activity [56].
- Aliquoting Protocol: Upon receipt, immediately aliquot the TUBE reagent into single-use volumes using low-binding tubes to minimize adsorption losses [56]. This practice is critical to avoid repeated freeze-thaw cycles, which can denature proteins and cause irreversible activity loss [56]. Aliquoting also safeguards the main stock from potential contamination during routine use.
Protection from Light and Moisture
TUBE reagents are susceptible to environmental degradation.
- Light Sensitivity: Conjugated TUBEs, especially those with fluorescent tags, are highly sensitive to photobleaching. Always store them in amber vials or tubes wrapped in aluminum foil to prevent light exposure, even during short-term storage in freezers or refrigerators [56].
- Moisture Control: For reagents stored at 2–8°C, ensure containers are tightly sealed. For sensitive lyophilized powders or materials in humid environments, consider storing reagents in a sealed container with desiccants to maintain stability [57].
Inventory Management
Implement a robust inventory system to track reagent usage and viability.
- FIFO Principle: Adopt a "First-In, First-Out" approach to ensure older stock is used before newer shipments [58].
- Digital Tracking: Label all aliquots with the reagent name, concentration, date received, date aliquoted, and expiration date [57]. Using a digital inventory system with barcodes can enhance traceability and simplify stock management [57].
- Regular Audits: Conduct monthly inventory inspections to identify and dispose of expired reagents, maintaining the integrity of your research [57].
Table 1: Summary of Storage Conditions for TUBE Reagents
| Parameter | Specification | Rationale |
|---|---|---|
| Unconjugated TUBEs | –20°C or –80°C | Long-term protein stability [56] |
| Conjugated TUBEs | 2–8°C (Refrigerated) | Prevents damage to the conjugate [56] |
| Aliquoting | Mandatory, single-use volumes | Prevents freeze-thaw damage and contamination [56] |
| Light Protection | Amber vials or foil wrapping | Prevents photobleaching of conjugates [56] |
| Container Type | Low-binding tubes | Minimizes protein adsorption to tube walls [56] |
| Inventory Control | FIFO system, digital logging | Prevents use of expired or degraded reagents [57] [58] |
Experimental Protocol: Enrichment of Ubiquitinated Proteins Using TUBEs
The following protocol details the tandem enrichment of ubiquitinated peptides using a TUBE-based approach, adapted from the SCASP-PTM methodology for use with TUBE reagents [49]. This workflow is designed for mass spectrometric analysis and can be modified for western blotting.
Workflow: TUBE-Based Ubiquitinated Protein Enrichment
Materials and Reagents
- Research Reagent Solutions:
- Lysis Buffer: A modified RIPA buffer, supplemented with 1-2% SDS, protease inhibitors, and 10-20 mM N-Ethylmaleimide (NEM) or Iodoacetamide (IAA) to inhibit deubiquitinating enzymes (DUBs) and preserve ubiquitination signals [4].
- TUBE Reagent: Agarose- or magnetic bead-conjugated TUBEs for affinity purification.
- Wash Buffers: High-salt buffers (e.g., with 500 mM NaCl) and low-detergent buffers to remove non-specifically bound proteins.
- Elution Buffer: A solution containing 2-4% SDS or a low-pH buffer (e.g., 0.1-0.2 M Glycine, pH 2.5-3.0) to efficiently dissociate the ubiquitin-TUBE complex.
- Trypsin: Sequencing-grade trypsin for protein digestion.
Step-by-Step Procedure
Protein Extraction and Denaturation:
- Lyse cells or tissue in pre-chilled SDS-containing lysis buffer. The SDS ensures efficient extraction and denaturation of proteins, inactivating DUBs.
- Heat the lysate at 95°C for 5-10 minutes to fully denature proteins and eliminate DUB activity.
- Clarify the lysate by centrifugation at >14,000 x g for 15 minutes at 4°C. Transfer the supernatant to a new tube [4].
Enrichment of Ubiquitinated Proteins:
- Dilute the clarified lysate 1:10 with a no-SDS or low-SDS buffer to reduce the SDS concentration below 0.1% for compatibility with the TUBE reagent.
- Incubate the diluted lysate with the TUBE-conjugated beads for 2-4 hours at 4°C with gentle rotation [4].
Washing:
- Pellet the beads (by centrifugation or magnet) and carefully remove the supernatant.
- Wash the beads 3-4 times with 1 mL of wash buffer (e.g., PBS with 500 mM NaCl and 0.1% Tween-20) to remove non-specifically bound proteins [4].
Elution and Digestion:
Cleanup and Analysis:
Table 2: Key Reagent Solutions for TUBE-Based Enrichment
| Reagent | Function | Critical Components & Notes |
|---|---|---|
| Lysis Buffer | Extracts and denatures proteins, inhibits DUBs. | SDS (1-2%), Protease Inhibitors, NEM (10-20 mM) [4]. |
| TUBE Reagent | Binds and enriches poly-ubiquitinated proteins. | Agarose/magnetic bead-conjugated; linkage-specific or pan-specific available [4]. |
| Wash Buffer | Removes non-specifically bound proteins. | High-salt (500 mM NaCl), low-detergent (0.1% Tween-20) [4]. |
| Elution Buffer | Releases enriched proteins from TUBE reagent. | SDS (2-4%) or low-pH Glycine buffer; compatible with downstream MS [4]. |
| Trypsin | Digests enriched proteins into peptides for MS. | Sequencing-grade for efficient and specific digestion [4]. |
The Scientist's Toolkit: Essential Materials
A successful TUBE experiment requires more than just the core reagent. The table below lists essential materials and their functions.
Table 3: Essential Research Reagent Solutions and Materials
| Item | Function | Specification Notes |
|---|---|---|
| Low-Binding Tubes | Aliquoting & sample handling. | Polypropylene; pre-labeled for tracking [56]. |
| Temperature Monitoring Device | Monitors storage units. | Calibrated data logger for –80°C/–20°C/4°C [58]. |
| Magnetic/Agarose Beads | Solid support for TUBE immobilization. | Choice depends on lab preference and equipment [4]. |
| SDS (Sodium Dodecyl Sulfate) | Strong denaturant in lysis buffer. | Ensures complete protein extraction and DUB inhibition [4]. |
| Deubiquitinase (DUB) Inhibitors | Preserves ubiquitin signature. | N-Ethylmaleimide (NEM) or Iodoacetamide (IAA) [4]. |
| Mass Spectrometry-Grade Trypsin | Digests proteins for LC-MS/MS. | Ensves high cleavage efficiency and low autolysis [4]. |
| C18 Desalting Columns | Desalts and cleans peptide mixtures. | Essential for sample preparation prior to LC-MS/MS [49]. |
| Linkage-Specific Ub Antibodies | Validation of enrichment (e.g., by WB). | Antibodies for K48, K63, etc., to confirm results [4]. |
Benchmarking TUBE Technology: A Critical Comparison with Alternative Methods
Within the ubiquitin-proteasome system, the precise enrichment of polyubiquitinated proteins is a foundational step for deciphering the roles of ubiquitination in cellular regulation and disease. For years, the field has relied heavily on ubiquitin antibodies for this task. However, the emergence of Tandem Ubiquitin Binding Entities (TUBEs) represents a significant technological advance. This application note provides a direct comparison of the affinity and specificity of TUBEs versus traditional ubiquitin antibodies, framing the discussion within the context of enriching ubiquitinated proteins for downstream analysis. We include quantitative data, detailed protocols for key applications, and visual workflows to guide researchers in selecting the optimal tool for their experimental goals.
Technology Comparison: TUBEs vs. Traditional Antibodies
The core difference between these tools lies in their design and mechanism of action. Traditional ubiquitin antibodies are immunoglobulin-based reagents that recognize specific epitopes on ubiquitin. In contrast, TUBEs are engineered, tandem-repeated ubiquitin-binding domains (UBDs) that harness avidity to achieve high-affinity binding to polyubiquitin chains [6] [16].
Table 1: Direct Comparison of TUBEs and Traditional Ubiquitin Antibodies
| Feature | TUBEs | Traditional Ubiquitin Antibodies |
|---|---|---|
| Affinity (Kd) | Nanomolar range (1-10 nM) for polyubiquitin chains [6] | Variable; often micromolar or weaker, leading to lower capture efficiency [6] [16] |
| Specificity | High specificity for polyubiquitin chains; chain-selective variants available (e.g., K48, K63, M1) [6] [25] | Notoriously non-selective; pan-specific antibodies can bind mono-ubiquitin and unrelated proteins, causing artifacts [6] |
| Polyubiquitin Chain Protection | Yes, protect chains from deubiquitinases (DUBs) and proteasomal degradation, even without inhibitors [6] | No, offer no inherent protection during lysis and processing. |
| Primary Applications | Pulldown/MS, HTS assays, Western blot (alternative to antibodies), cellular imaging [6] [25] [5] | Immunoprecipitation (IP), Western blot, immunohistochemistry [16] |
| Key Advantage | High-affinity capture, linkage-specific analysis, and preservation of ubiquitination state. | Wide historical use and familiarity; direct compatibility with many established protocols. |
The quantitative data in Table 1 highlights the superior affinity of TUBEs, which bind polyubiquitin in the nanomolar range [6]. This is a critical improvement over the weaker and less selective binding often observed with traditional ubiquitin antibodies, which can lead to co-enrichment of non-ubiquitinated proteins and high background [6] [16]. Furthermore, TUBEs offer a unique functional advantage: they act as guardians of the ubiquitin code by shielding polyubiquitin chains from deubiquitinating enzymes and proteasomal degradation during cell lysis and processing, maintaining the native ubiquitination state even in the absence of enzyme inhibitors [6].
Experimental Protocols
Below are detailed methodologies for leveraging TUBEs in two key applications: the enrichment of ubiquitinated proteins for mass spectrometry and a high-throughput assay for linkage-specific ubiquitination.
Protocol 1: Enrichment of Ubiquitinated Proteins using TUBE Pulldown for Mass Spectrometry
This protocol describes a procedure for using TUBE agarose beads (e.g., LifeSensors UM501M) to isolate ubiquitinated proteins from cell lysates for subsequent proteomic analysis [6].
Key Research Reagent Solutions:
- TUBE Agarose Beads: Tandem ubiquitin-binding entities immobilized on agarose resin for affinity purification.
- Lysis Buffer: Must be optimized to preserve polyubiquitination. A typical formulation includes 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, supplemented with protease inhibitors and, optionally, DUB inhibitors (though TUBEs offer inherent protection) [25].
- Wash Buffer: Similar to lysis buffer but may have a reduced detergent concentration (e.g., 0.1% NP-40).
- Elution Buffer: A solution containing high concentrations of free ubiquitin (e.g., 1-2 mg/mL) to competitively elute bound proteins, or a low-pH buffer like 0.1 M glycine (pH 2.5-3.0).
Step-by-Step Procedure:
- Cell Lysis: Harvest and lyse cells in an appropriate volume of pre-chilled lysis buffer. Incubate on ice for 15-30 minutes.
- Clarification: Centrifuge the lysate at >15,000 × g for 15 minutes at 4°C. Transfer the supernatant (soluble fraction) to a new tube.
- Protein Quantification: Determine the protein concentration of the lysate using a standard assay (e.g., BCA).
- Pre-Clearance (Optional): Incubate the lysate with control agarose beads for 30-60 minutes to reduce non-specific binding. Centrifuge and collect the supernatant.
- TUBE Incubation: Add 20-50 µL of TUBE agarose bead slurry to 500 µg - 1 mg of total protein. Adjust the final volume with lysis buffer if necessary. Incubate for 2-4 hours at 4°C with end-over-end mixing.
- Washing: Pellet the beads by gentle centrifugation. Carefully remove the supernatant and wash the beads 3-4 times with 1 mL of wash buffer.
- Elution: Elute the bound ubiquitinated proteins by incubating the beads with 2-3 bead volumes of elution buffer for 5-10 minutes at room temperature. Repeat the elution once and pool the eluates.
- Neutralization: If using low-pH elution, immediately neutralize the pooled eluates with a neutralization buffer (e.g., 1 M Tris-HCl, pH 8.0).
- Downstream Analysis: The eluted proteins can now be processed for mass spectrometry analysis (e.g., tryptic digestion, desalting) or analyzed by Western blot.
The following workflow diagram illustrates the key steps of the TUBE pulldown protocol:
Protocol 2: HTS Assay for Linkage-Specific Ubiquitination using TUBEs
This protocol leverages chain-selective TUBEs (e.g., K48 or K63 TUBEs) immobilized on a microtiter plate to capture and quantify the linkage-specific ubiquitination of an endogenous target protein, such as RIPK2, in a high-throughput format [25].
Key Research Reagent Solutions:
- Chain-Selective TUBEs: TUBEs specific for K48 or K63 linkages, biotinylated for immobilization on streptavidin plates.
- Coating Buffer: Phosphate-Buffered Saline (PBS), pH 7.4.
- Assay Diluent/Blocking Buffer: PBS containing 1-3% Bovine Serum Albumin (BSA) or 5% non-fat dry milk.
- Cell Lysis Buffer: As described in Protocol 3.1.
- Primary Antibody: A highly specific antibody against the protein of interest (e.g., anti-RIPK2).
- Detection Antibody: An HRP-conjugated secondary antibody.
Step-by-Step Procedure:
- Plate Coating: Coat a streptavidin-coated microtiter plate with biotinylated chain-selective TUBEs (e.g., 1-2 µg/mL in PBS) for 1-2 hours at room temperature or overnight at 4°C.
- Blocking: Aspirate the coating solution and block the wells with 200-300 µL of blocking buffer for 1-2 hours at room temperature to prevent non-specific binding.
- Cell Treatment and Lysis: Treat cells (e.g., THP-1 monocytes) with the desired stimulus (e.g., L18-MDP to induce K63-ubiquitination of RIPK2) or PROTAC (e.g., RIPK2 degrader to induce K48-ubiquitination). Lyse the cells and clarify the lysates as in Protocol 3.1.
- Sample Incubation: Add the clarified cell lysates to the TUBE-coated wells. Incubate for 2-3 hours at room temperature with gentle shaking to allow capture of ubiquitinated proteins.
- Washing: Wash the plate 3-5 times with a wash buffer (e.g., PBS with 0.05% Tween-20).
- Target Detection: Incubate with a primary antibody against the target protein (e.g., anti-RIPK2) for 1-2 hours, followed by an HRP-conjugated secondary antibody for 1 hour.
- Signal Development and Readout: Develop the signal using a chemiluminescent or colorimetric HRP substrate and read on a plate reader.
The application of chain-selective TUBEs in a high-throughput screening (HTS) context allows for the dissection of complex ubiquitination dynamics, as summarized below:
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagents for TUBE-Based Ubiquitin Research
| Reagent | Function | Example Application |
|---|---|---|
| Pan-Selective TUBEs | Broadly captures all types of polyubiquitin chains with high affinity. | General enrichment of the total ubiquitinated proteome for pulldown/MS [6] [5]. |
| K48-Selective TUBEs | Specifically enriches K48-linked polyubiquitin chains, the primary signal for proteasomal degradation. | Studying PROTAC-mediated target ubiquitination and degradation [6] [25]. |
| K63-Selective TUBEs | Specifically enriches K63-linked chains, key regulators of signal transduction and inflammation. | Investigating inflammatory signaling pathways (e.g., NF-κB activation) [25]. |
| TAMRA-Labeled TUBEs | Fluorescently tagged TUBEs for imaging ubiquitin dynamics in fixed or live cells. | Visualizing the spatial and temporal distribution of polyubiquitin chains [6]. |
| DUB Inhibitors | Chemical inhibitors of deubiquitinating enzymes (e.g., PR-619). | Used in conjunction with or without TUBEs to further stabilize ubiquitin signals in lysates. |
The direct comparison of TUBEs and traditional ubiquitin antibodies clearly demonstrates that TUBEs offer a paradigm shift in the enrichment and study of ubiquitinated proteins. Their superior nanomolar affinity, enhanced specificity—particularly with chain-selective variants—and unique ability to protect polyubiquitin chains from degradation make them an indispensable tool for modern ubiquitin research. By implementing the detailed application notes and protocols provided here, researchers can more effectively decipher the complex language of the ubiquitin code, accelerating discovery in basic biology and drug development, particularly in the burgeoning field of targeted protein degradation.
The comprehensive analysis of the ubiquitin-modified proteome, or ubiquitinome, is critical for understanding its extensive role in cellular regulation and disease. Two powerful methodologies have emerged as cornerstones in this endeavor: Tandem-repeated Ubiquitin-Binding Entities (TUBEs) and diGly remnant antibody-based affinity enrichment. Used independently, each method provides valuable insights; however, their strategic integration creates a synergistic workflow that leverages the unique advantages of both to achieve unprecedented depth and accuracy in ubiquitinome mapping. This application note details the underlying principles, practical protocols, and analytical benefits of combining TUBE-based protein-level enrichment with diGly antibody-based peptide-level enrichment for mass spectrometric analysis, providing a robust framework for researchers and drug development professionals.
Table 1: Core Characteristics of Ubiquitin Enrichment Techniques
| Feature | TUBE Enrichment | diGly Antibody Enrichment |
|---|---|---|
| Enrichment Level | Protein-level | Peptide-level |
| Primary Target | Polyubiquitin chains of various linkages [4] | K-ε-GG remnant on tryptic peptides [59] |
| Key Advantage | Protects ubiquitin chains from deubiquitinases (DUBs) and preserves endogenous chain architecture [4] | Provides precise, site-specific identification of ubiquitination sites [59] [60] |
| Linkage Information | Can be retained when using linkage-specific TUBEs [4] | Typically lost during tryptic digestion (collapses chain architecture) [61] |
| Compatibility | Ideal for initial capture from complex lysates; compatible with any downstream analysis [4] | Requires tryptic digestion prior to enrichment; ideal for LC-MS/MS [59] |
The Scientist's Toolkit: Essential Research Reagents
Successful execution of the combined TUBE-diGly protocol requires specific, high-quality reagents. The following table details the essential components and their critical functions within the workflow.
Table 2: Key Research Reagent Solutions for Combined Ubiquitin Enrichment
| Reagent / Kit | Function / Principle | Application Note |
|---|---|---|
| TUBE Reagents (e.g., Agarose-conjugated TUBEs) | Protein-level enrichment; tandem ubiquitin-binding domains with high affinity for polyUb chains, offering protection from DUBs [4]. | Select generic TUBEs for total ubiquitinome, or linkage-specific TUBEs (e.g., K48- or K63-specific) for targeted studies [4]. |
| diGLY Antibody (e.g., PTMScan Ubiquitin Remnant Motif Kit) | Peptide-level immunoaffinity enrichment; highly specific antibody recognizing the K-ε-GG remnant left on ubiquitinated peptides after trypsin digestion [59] [60]. | The cornerstone for site-specific identification; critical for mass spectrometry analysis. |
| Cell Lysis Buffer (with inhibitors) | 8M Urea, 50mM Tris-HCl (pH 8), 150mM NaCl, Protease Inhibitors, 5mM N-Ethylmaleimide (NEM), 1mM Na-Orthovanadate [59] [62]. | Urea denatures proteins; NEM alkylates cysteine and inhibits DUBs [59]; Orthovanadate improves tyrosine phosphoprotein and ubiquitin enrichment [62]. |
| Siliconized / Low-Bind Tubes | Laboratory consumables with a treated surface to minimize non-specific binding of proteins and peptides. | Essential throughout the protocol to prevent loss of low-abundance ubiquitinated species. |
Quantitative Superiority of the Integrated Workflow
The sequential application of TUBE and diGly enrichment strategies yields a significant multiplicative effect on analytical performance. The data-independent acquisition (DIA) mass spectrometry method, when applied to diGly-enriched peptides, has been shown to identify over 35,000 distinct diGly peptides in a single measurement, doubling the number and quantitative accuracy achieved by traditional data-dependent acquisition (DDA) [60]. This integrated approach provides the sensitivity and reproducibility required for systems-wide studies of ubiquitin signaling.
Table 3: Performance Metrics of Advanced diGly Proteomics
| Metric | Standard diGly (DDA) | Advanced diGly (DIA) | Key Parameter |
|---|---|---|---|
| DiGly Peptides (Single Shot) | ~20,000 [60] | ~35,000 [60] | Proteasome-inhibited HEK293 cells |
| Quantitative Reproducibility (CV < 20%) | 15% of peptides [60] | 45% of peptides [60] | Coefficient of variation (CV) across replicates |
| Total Unique DiGly Sites | N/A | 89,650 (from consolidated libraries) [60] | From HEK293, U2OS, and untreated cell libraries |
| Sample Input Post-Enrichment | 100% | 25% [60] | Amount of enriched material injected for LC-MS/MS |
Detailed Experimental Protocol
Stage 1: TUBE-Based Enrichment of Ubiquitinated Proteins
Goal: To isolate and protect ubiquitinated proteins from complex lysates.
Procedure:
- Cell Lysis: Harvest cells or tissue and lyse in a pre-chilled, high-denaturant lysis buffer (8M Urea, 50mM Tris-HCl pH 8, 150mM NaCl) supplemented with complete protease inhibitors, 5mM NEM, and 1mM Sodium Orthovanadate [59] [62]. Note: NEM is critical to inhibit deubiquitinases (DUBs) and preserve ubiquitin signals [59].
- Clarification: Centrifuge lysates at 20,000 x g for 15 minutes at 4°C. Transfer the supernatant to a new siliconized tube.
- Protein Quantification: Determine protein concentration using a compatible assay (e.g., BCA).
- TUBE Incubation: Incubate 1-5 mg of protein lysate with 20-50 µL of agarose-conjugated TUBE beads for 2-4 hours at 4°C with end-over-end mixing.
- Washing: Pellet beads via gentle centrifugation and wash sequentially with:
- 3x with 10 bead volumes of lysis buffer.
- 2x with 10 bead volumes of 50mM Ammonium Bicarbonate (ABC) pH 8.0.
- On-Bead Digestion (or Elution): Proceed directly to on-bead tryptic digestion (Stage 2). Alternatively, ubiquitinated proteins can be eluted using standard SDS-PAGE sample buffer for western blotting or other analyses.
Stage 2: Protein Digestion and Peptide Cleanup
Goal: To generate peptides containing the diGly remnant for subsequent immunoaffinity purification.
Procedure:
- Reduction and Alkylation: Resuspend TUBE beads in 50mM ABC. Add dithiothreitol (DTT) to 5mM and incubate at 37°C for 30 minutes to reduce disulfide bonds. Then add iodoacetamide (IAA) to 15mM and incubate at room temperature in the dark for 30 minutes to alkylate cysteines.
- Protein Digestion:
- Peptide Desalting: Acidify the digest with 1% Trifluoroacetic Acid (TFA). Desalt the peptides using a C18 solid-phase extraction (SPE) cartridge or SepPak column [59]. Elute peptides with 50% Acetonitrile (ACN), 0.5% Acetic Acid (HAcO). Lyophilize the eluate to dryness.
Stage 3: diGly Peptide Immunoaffinity Enrichment and MS Analysis
Goal: To specifically isolate K-ε-GG-containing peptides for high-sensitivity mass spectrometry.
Procedure:
- Peptide Reconstitution: Reconstitute the dried peptide pellet in 1.4 mL of Immunoaffinity Purification (IAP) buffer (50mM MOPS-NaOH pH 7.2, 10mM Na2HPO4, 50mM NaCl).
- diGly Antibody Incubation: Add the recommended amount of anti-K-ε-GG antibody (e.g., 31.25 µg antibody per 1 mg peptide input) [60] and incubate for 2 hours at 4°C with rotation.
- Washing and Elution:
- StageTip Cleanup: Desalt the eluted diGly peptides using C18 StageTips to remove salts and residual TFA before MS analysis.
- LC-MS/MS Analysis:
- Chromatography: Separate peptides on a reverse-phase C18 column using a nanoflow LC system.
- Mass Spectrometry: Analyze using an Orbitrap-based mass spectrometer. DIA (Data-Independent Acquisition) is highly recommended over DDA for its superior quantitative accuracy, sensitivity, and data completeness in diGly analyses [60]. The optimized DIA method should use ~46 precursor isolation windows and high MS2 resolution (30,000) [60].
Critical Technical Considerations and Troubleshooting
Specificity Note: The diGly antibody also enriches peptides modified by the ubiquitin-like proteins NEDD8 and ISG15, which generate an identical K-ε-GG remnant. In most human cell studies, these modifications account for less than 6% of identified diGly peptides [60]. For exclusive analysis of ubiquitination, consider using a longer remnant antibody after LysC digestion [4] [61].
Fractionation for Depth: For maximal ubiquitinome depth, implement a fractionation step prior to diGly enrichment. A simple, fast offline high-pH reverse-phase fractionation of tryptic peptides into just three fractions can dramatically increase diGly peptide identifications, enabling the detection of over 23,000 sites from a single sample without prohibitive complexity [63].
Handling Abundant Peptides: In proteasome-inhibited samples, K48-linked ubiquitin-chain-derived diGly peptides become extremely abundant and can dominate the signal. Separating these highly abundant peptides via pre-fractionation before diGly enrichment prevents them from competing for antibody binding sites and masking lower-abundance peptides, thus improving overall coverage [60].
Comparative Analysis of Commercially Available TUBE Platforms (e.g., LifeSensors, Cell Signaling Technology)
Tandem Ubiquitin Binding Entities (TUBEs) represent a transformative technology in the field of ubiquitin proteomics, specifically designed to address long-standing challenges in the enrichment and analysis of polyubiquitinated proteins. Traditional methods for studying protein ubiquitination have relied heavily on immunoprecipitation of overexpressed epitope-tagged ubiquitin or the use of ubiquitin antibodies, which are often plagued by issues of low affinity, poor selectivity, and significant artifacts [6]. TUBE technology circumvents these limitations by harnessing the combined strength of multiple Ubiquitin Binding Domains (UBDs) arranged in tandem, resulting in affinities for polyubiquitin chains in the nanomolar range (Kd 1-10 nM) [6]. This architectural innovation provides up to a 1000-fold increase in affinity for polyubiquitin moieties compared to single UBA domains, enabling more reliable capture and detection of ubiquitinated proteins even at low abundance levels [64].
The significance of TUBE technology extends across multiple research domains, particularly in drug discovery and development, where understanding the ubiquitin-proteasome system is crucial for advancing targeted protein degradation (TPD) strategies such as PROTACs (Proteolysis Targeting Chimeras) and molecular glues [6]. As pharmaceutical research increasingly focuses on harnessing cellular degradation machinery for therapeutic purposes, the ability to accurately monitor ubiquitination events becomes paramount. TUBEs serve as essential tools in this context, enabling researchers to quickly distinguish true hits from false positives, develop structure-activity relationships, and establish rank order potency from purified enzymes to cellular models [6]. Furthermore, the integration of TUBE-based approaches with advanced proteomic methods like mass spectrometry has opened new avenues for comprehensive ubiquitome analysis, providing unprecedented insights into the complex landscape of protein modification and degradation in cellular systems [65].
Comparative Analysis of Commercial TUBE Platforms
LifeSensors TUBE Portfolio
Table 1: LifeSensors TUBE Products for Ubiquitinated Protein Enrichment and Detection
| Product Name | Specificity | Tag/Conjugate | Applications | Quantity & Price |
|---|---|---|---|---|
| UM101: TUBE 1 | Pan-selective | GST | Pull down of polyubiquitylated proteins; Protection from deubiquitylation and degradation | 200µg - 1mg; $633 - $2,069 |
| UM301: TUBE 1 | Pan-selective | Biotin | Detection and enrichment | Not specified; $755 |
| UM302: TUBE 2 | Pan-selective | Biotin | Detection and enrichment | Not specified; $824 |
| UM304: K63 TUBE | K63-linkage specific | Biotin | Specific detection and enrichment of K63-linked chains | Not specified; $808 |
| UM307: K48 TUBE HF | K48-linkage specific | Biotin | Specific detection and enrichment of K48-linked chains | Not specified; $679 - $1,888 |
| UM306: M1 Linear TUBE | M1-linear linkage specific | Biotin | Specific detection and enrichment of linear ubiquitin chains | Not specified; $707 |
| UM312: TUBE 2 | Pan-selective | HRP | Detection applications | Not specified; $648 |
| UM502T | Pan-selective (TUBE 2) | TAMRA fluorophore | Imaging techniques | Not specified |
| UM401 | Pan-selective | Agarose-coupled | Pull down assays | Not specified |
LifeSensors offers the most comprehensive portfolio of TUBE reagents currently available commercially, with products spanning pan-selective and linkage-specific configurations for diverse research applications. Their pan-selective TUBEs (TUBE 1 and TUBE 2) exhibit broad specificity for all types of polyubiquitin chains and are available with various tags including GST, biotin, HRP, and fluorescent conjugates [66] [64]. These reagents demonstrate remarkable affinity for polyubiquitin chains, with Kd values in the 1-10 nM range, enabling highly efficient capture of ubiquitinated proteins even without the use of proteasome or deubiquitylase inhibitors that are normally required to prevent the rapid turnover of polyubiquitin signals in cell lysates [6]. The protective function of LifeSensors TUBEs represents a significant advantage over traditional methods, as they shield polyubiquitinated proteins from both deubiquitinating enzymes and proteasome-mediated degradation during processing [64].
Beyond their pan-selective offerings, LifeSensors has developed specialized linkage-specific TUBEs that target particular ubiquitin chain architectures, including K48-linked, K63-linked, and M1-linear (methionine-1 linked) chains [66]. These linkage-specific reagents are particularly valuable for investigating the distinct biological functions associated with different ubiquitin chain types. For instance, K48-linked chains primarily target proteins for proteasomal degradation, while K63-linked chains typically mediate non-proteolytic signaling events in processes such as DNA repair, inflammation, and protein trafficking [65]. The K48 TUBE HF (High Fidelity) and K63 TUBE products enable researchers to selectively enrich and study these functionally distinct ubiquitin signatures, providing crucial insights into the specific ubiquitin-dependent pathways operating in their experimental systems [66].
Cell Signaling Technology Platforms
The search results reveal that Cell Signaling Technology (CST) offers magnetic separation racks designed for immunoprecipitation procedures but do not indicate the availability of specific TUBE products for ubiquitin enrichment [67] [68]. The 12-Tube Magnetic Separation Rack (#14654) and 6-Tube Magnetic Separation Rack (#7017) are designed for small-scale isolation of immunocomplexes using magnetic beads and can be used with various Protein A, Protein G, and ChIP-Grade Protein G Magnetic Beads [67] [68]. While these tools could potentially support TUBE-based workflows when combined with appropriate magnetic bead-conjugated ubiquitin binding reagents, the absence of specific TUBE products in CST's portfolio suggests that LifeSensors currently maintains a specialized focus on tandem ubiquitin binding entity technology.
Table 2: Comparison of Platform Specialization and Capabilities
| Feature | LifeSensors | Cell Signaling Technology |
|---|---|---|
| Core TUBE Offerings | Comprehensive portfolio (pan-selective and linkage-specific) | No specific TUBE products identified |
| Technology Specialization | Tandem Ubiquitin Binding Entities | Antibodies, magnetic separation equipment |
| Key Applications | Ubiquitin proteomics, PROTAC development, protein degradation studies | General protein analysis, chromatin immunoprecipitation |
| Supporting Tools | TUBE-based mass spectrometry services, assay development | Magnetic beads, separation racks |
| Price Range | ~$633-$2,069 for standard TUBEs | ~Not applicable |
Essential Research Reagent Solutions for TUBE-Based Research
Table 3: Key Research Reagent Solutions for TUBE Experiments
| Reagent/Tool | Function/Application | Example Products |
|---|---|---|
| Pan-Selective TUBEs | Broad enrichment of all polyubiquitin chain types | LifeSensors UM101 (GST), UM301 (Biotin), UM302 (Biotin) |
| Linkage-Specific TUBEs | Selective enrichment of specific ubiquitin chain linkages | LifeSensors K48 TUBE HF, K63 TUBE, M1 Linear TUBE |
| Detection TUBEs | Direct detection of ubiquitinated proteins via conjugated reporters | LifeSensors UM312 (HRP), UM502T (TAMRA) |
| Magnetic Separation Systems | Isolation of bead-bound complexes | CST 12-Tube Magnetic Separation Rack (#14654) |
| Cell Culture Platforms | Advanced in vitro models for drug screening | Microfluidics-based 2D/3D culture systems [69] |
| Mass Spectrometry Platforms | Identification and quantification of ubiquitinated proteins | LifeSensors TUBE-based proteomics services [65] |
A successful TUBE-based research program requires careful selection of complementary reagents and tools that support the entire experimental workflow from sample preparation to data analysis. The core TUBE reagents themselves represent the foundation, with pan-selective variants being ideal for global ubiquitome profiling and linkage-specific versions enabling targeted investigation of particular ubiquitin signaling pathways [66] [6]. The choice of tag or conjugate—GST for pull-down assays, biotin for streptavidin-based detection or capture, HRP for direct chemiluminescent detection, or fluorophores like TAMRA for imaging applications—should align with the intended experimental endpoint and detection methodology [66] [64].
Magnetic separation systems, such as those offered by Cell Signaling Technology, provide practical laboratory infrastructure for efficient processing of TUBE-bound complexes when using magnetic bead-conjugated versions of the reagents [67] [68]. These racks are designed specifically for small-scale isolation of immunocomplexes from volumes of 1.5-2.0 ml, featuring powerful neodymium rare earth magnets that enable rapid clearance of solutions (typically within 1-2 minutes) and gentle resuspension of beads for washing and elution steps [67]. For cell-based studies, particularly in drug discovery applications, advanced cell culture platforms including microfluidics-based systems that better mimic in vivo conditions can enhance the biological relevance of TUBE experiments by providing more physiologically accurate cellular contexts [69].
Finally, mass spectrometry platforms represent crucial components for comprehensive ubiquitome analysis, with LifeSensors offering specialized TUBE-based proteomics services that combine their enrichment technology with advanced LC-MS/MS capabilities [65]. This integrated approach enables both qualitative and quantitative assessment of ubiquitinated proteins, including identification of specific ubiquitination sites and chain linkage types, providing researchers with detailed insights into ubiquitin-dependent regulatory mechanisms in their experimental systems.
Detailed Experimental Protocols for TUBE Applications
Protocol 1: Enrichment of Polyubiquitinated Proteins from Cell Lysates Using GST-Tagged TUBEs
Principle: This protocol describes the procedure for isolating polyubiquitinated proteins from cell lysates using GST-tagged TUBEs (e.g., LifeSensors UM101), which can be coupled with glutathione resin for pull-down assays. The method leverages the high affinity of TUBEs for polyubiquitin chains while protecting them from deubiquitinating enzymes and proteasomal degradation during processing.
Reagents and Equipment:
- LifeSensors UM101: TUBE 1 (GST) [64]
- Cell lysis buffer (recommended: 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 1 mM EDTA) supplemented with fresh protease inhibitors
- Glutathione resin or glutathione-sepharose beads
- Wash buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% NP-40)
- Elution buffer (50 mM Tris-HCl, pH 8.0, 20 mM reduced glutathione)
- Benchtop centrifuge
- Rotating mixer or end-over-end rotator
Procedure:
- Cell Lysis and Preparation: Harvest cells and lyse using appropriate lysis buffer. Maintain samples at 4°C throughout the procedure. Centrifuge lysates at 14,000 × g for 15 minutes at 4°C to remove insoluble debris. Transfer the supernatant to a fresh tube and determine protein concentration.
- TUBE-Binding Incubation: Add 2-5 µg of GST-tagged TUBE (UM101) per 500 µg of total cell lysate protein. Incubate for 2 hours at 4°C with gentle rotation.
- Bead Capture: Add pre-washed glutathione resin (20-50 µl slurry per reaction) to the lysate-TUBE mixture. Continue incubation for an additional 1-2 hours at 4°C with rotation.
- Washing: Pellet beads by brief centrifugation (500 × g for 2 minutes) and carefully remove supernatant. Wash beads three times with 1 ml wash buffer, resuspending completely with each wash.
- Elution: After final wash, completely remove wash buffer and elute bound proteins by adding 40-100 µl elution buffer. Incubate for 10 minutes at room temperature with gentle mixing. Pellet beads and collect supernatant containing enriched polyubiquitinated proteins.
- Analysis: Analyze eluates by Western blotting using ubiquitin-specific antibodies, or process for mass spectrometry analysis.
Technical Notes:
- For proteomics applications, include proteasome inhibitors (e.g., MG132) and deubiquitinase inhibitors (e.g., PR-619) in lysis buffer when not using TUBEs, though TUBEs themselves provide substantial protection [6] [64].
- Optimization of TUBE:lysate ratio may be necessary for different sample types.
- For downstream mass spectrometry, elution can be performed using SDS-PAGE sample buffer followed by gel separation and in-gel digestion [65].
Protocol 2: TUBE-Based Mass Spectrometry Proteomics for Ubiquitome Analysis
Principle: This protocol outlines the procedure for large-scale identification of ubiquitinated proteins using TUBE enrichment followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The approach enables both qualitative identification and quantitative analysis of ubiquitination sites and chain linkage types.
Reagents and Equipment:
- LifeSensors pan-selective or linkage-specific TUBEs (biotinylated or GST-tagged) [66] [65]
- Streptavidin magnetic beads (if using biotinylated TUBEs) or glutathione resin (if using GST-tagged TUBEs)
- Strong denaturing lysis buffer (e.g., 6 M urea, 2 M thiourea in 50 mM Tris-HCl, pH 7.5)
- Reduction and alkylation reagents: DTT or TCEP; iodoacetamide
- Trypsin or other proteases for digestion
- Mass spectrometry-compatible solvents: 0.1% formic acid in water, 0.1% formic acid in acetonitrile
- LC-MS/MS system
Procedure:
- Sample Preparation and Lysis: Lyse cells in denaturing lysis buffer with sonication to ensure complete disruption. Clarify by centrifugation at 14,000 × g for 15 minutes.
- TUBE Enrichment: Incubate clarified lysate (1-5 mg total protein) with appropriate TUBEs (5-20 µg per mg lysate protein) for 2 hours at 4°C with rotation. Capture TUBE-protein complexes using appropriate beads (streptavidin for biotinylated TUBEs, glutathione resin for GST-tagged TUBEs).
- On-Bead Digestion: Wash beads extensively with lysis buffer followed by 50 mM ammonium bicarbonate buffer. Reduce disulfide bonds with 5 mM DTT (30 minutes, 37°C) and alkylate with 15 mM iodoacetamide (30 minutes, room temperature in dark). Digest proteins on-bead with trypsin (1:50 enzyme-to-protein ratio) overnight at 37°C.
- Peptide Cleanup: Acidify digests with 1% trifluoroacetic acid and desalt using C18 solid-phase extraction cartridges or StageTips.
- LC-MS/MS Analysis: Reconstitute peptides in 0.1% formic acid and analyze by nanoLC-MS/MS using a gradient of increasing acetonitrile.
- Data Analysis: Search MS/MS data against appropriate protein databases using search engines that can accommodate ubiquitin remnant peptides (e.g., with GlyGly modification on lysine residues). Use software tools to quantify changes in ubiquitination sites across conditions.
Technical Notes:
- LifeSensors offers specialized TUBE-based mass spectrometry services that typically deliver results within 2-4 weeks [65].
- For linkage type identification, monitor for signature peptides corresponding to specific lysine residues in ubiquitin (e.g., K48, K63) [65].
- Quantitative approaches can be implemented using isobaric labeling (TMT, iTRAQ) or label-free methods.
Figure 1: General workflow for TUBE-based enrichment of ubiquitinated proteins, showing key steps from cell lysis to downstream analysis options including Western blot and mass spectrometry.
Applications in Drug Development and Biotechnology
The integration of TUBE technology into drug development pipelines, particularly in the emerging field of targeted protein degradation, has created new opportunities for accelerating therapeutic discovery and validation. TUBEs serve as critical tools for monitoring the efficacy of PROTACs and molecular glues by enabling direct assessment of target protein ubiquitination, a key mechanistic step in induced protein degradation [6]. LifeSensors has developed high-throughput screening platforms that utilize TUBE technology to evaluate both polyubiquitylation and degradation of target proteins in plate-based formats, significantly accelerating PROTAC-based drug discovery efforts [6]. These approaches allow researchers to quickly distinguish true hits from false positives, develop structure-activity relationships, and establish rank order potency from purified enzymes to cellular models—all essential capabilities for efficient lead optimization in TPD programs.
Beyond early-stage discovery, TUBE-based approaches are finding applications in biomarker development and validation, particularly in neurodegenerative diseases where dysfunction of the ubiquitin-proteasome system is implicated in pathogenesis [6]. The ability to profile global ubiquitination signatures using TUBE-based mass spectrometry provides opportunities to identify disease-relevant ubiquitination patterns that could serve as diagnostic or prognostic indicators. LifeSensors has applied this approach in Alzheimer's disease research, receiving significant funding from the National Institutes of Health to develop PROTAC drug discovery and diagnostic platforms for this condition [66]. The combination of TUBE-based affinity enrichment with targeted mass spectrometry represents one of the most powerful methods for detecting alterations in post-translational modifications and identifying signatures with potential utility as research biomarkers or clinical diagnostics [6].
The regulatory landscape for drug development is also evolving to accommodate platform technologies that can streamline development processes. The FDA's Platform Technology Designation Program, established under section 506K of the FD&C Act, aims to create efficiencies in drug development, manufacturing, and review processes for drug product applications that incorporate designated platform technologies [70]. While TUBE technology itself would not typically be designated as a platform technology under this program, the therapeutic modalities it supports—such as PROTACs and other targeted protein degraders—could potentially benefit from such designation as they mature and demonstrate consistent performance across multiple product candidates.
The comparative analysis of commercially available TUBE platforms reveals that LifeSensors currently offers the most comprehensive and specialized portfolio of tandem ubiquitin binding entities, with products spanning pan-selective, linkage-specific, and detection-optimized configurations for diverse research applications. Their technology demonstrates significant advantages over traditional ubiquitin enrichment methods, including superior affinity for polyubiquitin chains (Kd 1-10 nM), protective functions that preserve ubiquitin signals during processing, and compatibility with multiple detection modalities and downstream analysis platforms [6] [64]. While Cell Signaling Technology provides supporting tools such as magnetic separation racks that can facilitate TUBE-based workflows, their portfolio does not appear to include specific TUBE reagents for ubiquitin enrichment [67] [68].
The application notes and protocols presented in this analysis provide researchers with practical frameworks for implementing TUBE-based approaches in their ubiquitin research programs, from basic enrichment and detection experiments to sophisticated proteomic profiling of the ubiquitome. As drug discovery efforts increasingly focus on targeted protein degradation and other ubiquitin-dependent processes, TUBE technology is poised to play an expanding role in validating therapeutic mechanisms, identifying biomarkers, and accelerating the development of novel therapeutics that harness the power of the ubiquitin-proteasome system. The continued refinement and application of these tools will undoubtedly yield new insights into ubiquitin biology and support the advancement of innovative treatment strategies for human diseases.
Within the framework of research focused on enriching ubiquitinated proteins using Tandem-repeated Ubiquitin-Binding Entities (TUBEs), validation of putative substrates and their specific ubiquitin chain linkages represents a critical step. The inherent complexity of the ubiquitin code, combined with the transient nature of ubiquitination and the activity of deubiquitinating enzymes (DUBs), makes reliable confirmation protocols essential [71] [72]. TUBEs function as powerful molecular traps, protecting polyubiquitinated proteins from both proteasomal degradation and deubiquitination, thereby enabling their isolation under native conditions [72]. However, the initial enrichment is only the beginning. Subsequent validation is required to confirm the identity of the ubiquitinated protein, the specific lysine residues modified, and the topology of the ubiquitin chain attached, as each linkage type—be it K48, K63, or others—can dictate entirely different functional outcomes for the substrate [71] [16]. These techniques are indispensable for researchers and drug development professionals aiming to decipher ubiquitination-related disease mechanisms and identify potential therapeutic targets.
Core Principles of Ubiquitination and Linkage Specificity
Ubiquitination is a reversible post-translational modification involving the covalent attachment of ubiquitin, a 76-amino acid protein, to substrate proteins. This process is mediated by a sequential enzymatic cascade comprising E1 (activating), E2 (conjugating), and E3 (ligating) enzymes [71] [16]. The modification can result in the attachment of a single ubiquitin molecule (monoubiquitination), multiple single ubiquitins (multi-monoubiquitination), or chains of ubiquitin (polyubiquitination) linked through one of its seven internal lysine residues (K6, K11, K27, K29, K33, K48, K63) or its N-terminal methionine (M1) [16].
The biological consequence of ubiquitination is predominantly determined by the linkage type of the polyubiquitin chain. For instance, K48-linked chains primarily target substrates for proteasomal degradation, whereas K63-linked chains are involved in non-proteolytic signaling pathways, such as DNA repair, kinase activation, and inflammation [71]. The specificity of these signals is further decoded by effector proteins containing ubiquitin-binding domains (UBDs) and is meticulously regulated by deubiquitinases (DUBs) that cleave ubiquitin chains with precise linkage preferences [73]. This intricate regulatory system maintains cellular homeostasis, and its dysregulation is implicated in numerous pathologies, including cancer and neurodegenerative diseases [71].
Table 1: Functions of Major Ubiquitin Linkage Types
| Linkage Type | Primary Functions |
|---|---|
| K48 | Targets substrates for proteasomal degradation [71] |
| K63 | Regulates protein-protein interactions, DNA repair, NF-κB signaling [71] |
| K11 | Cell cycle regulation, proteasomal degradation [71] |
| K6 | Mediates DNA damage repair [71] |
| K27 | Controls mitochondrial autophagy [71] |
| M1-linked | Regulates NF-κB inflammatory signaling [71] |
Validation Methodologies: From Confirmation to Linkage Mapping
Following the enrichment of ubiquitinated proteins using TUBEs, a multi-tiered validation strategy is required to confirm the modification and decipher the ubiquitin code. The techniques below are listed in a typical order of application, from general confirmation to highly specific linkage characterization.
Immunoblotting-Based Confirmation
The most straightforward and widely used method for initial validation is immunoblotting. This technique allows for the direct assessment of a protein's ubiquitination status after TUBE enrichment.
Detailed Protocol:
- Sample Preparation: Resuspend purified proteins isolated via TUBE pull-down in an appropriate loading buffer.
- Gel Electrophoresis: Separate the proteins by molecular weight using SDS-PAGE. The appearance of higher molecular weight smears or ladders above the expected size of the protein of interest is a classic indicator of polyubiquitination.
- Membrane Transfer: Transfer the separated proteins from the gel onto a nitrocellulose or PVDF membrane.
- Immunodetection: Probe the membrane with the following antibodies in a sequential manner:
- Primary Antibody: Use an antibody specific to your protein of interest. This confirms the presence of the substrate in the TUBE eluate.
- Primary Antibody for Ubiquitin: Re-probe the membrane with a general anti-ubiquitin antibody (e.g., P4D1, FK1/FK2) to confirm that the higher molecular weight species are indeed ubiquitinated.
- Linkage-Specific Antibodies: For linkage determination, use linkage-specific ubiquitin antibodies (e.g., specific for K48 or K63 chains). This step is crucial for inferring the potential function of the modification.
- Mutation Validation: To validate the ubiquitination site, perform the same workflow with cell lysates expressing a mutant form of the substrate protein where putative lysine residue(s) are replaced with arginine(s). A loss of the ubiquitination signal in the mutant confirms the site [16].
Advantages and Limitations:
- Advantages: Technically simple, widely accessible, and provides direct visual evidence of ubiquitination.
- Limitations: Low-throughput, does not identify specific modified lysine residues on the substrate, and linkage-specific antibodies can sometimes exhibit cross-reactivity [16].
Mass Spectrometry (MS) for Site-Specific Identification
For a comprehensive, high-resolution analysis of ubiquitination, mass spectrometry-based proteomics is the method of choice. It allows for the precise identification of the specific lysine residues on the substrate that are modified by ubiquitin and can also provide information on chain linkage.
Detailed Protocol:
- Enrichment: Isubiquitinated proteins using TUBEs from cell lysates under native conditions. This step protects the ubiquitin conjugates and reduces background [72].
- Denaturation and Digestion: Denature the enriched proteins and digest them into peptides using a protease like trypsin.
- Peptide Enrichment (Optional): Further enrich for ubiquitin-derived peptides using anti-di-glycine (anti-K-ε-GG) antibodies. This antibody specifically recognizes the diglycine remnant that is left attached to the modified lysine after tryptic digestion, serving as a mass tag for ubiquitination sites [16].
- LC-MS/MS Analysis: Separate the peptides by liquid chromatography (LC) and analyze them by tandem mass spectrometry (MS/MS).
- Data Analysis: Use bioinformatic tools to search the MS/MS spectra against protein databases. The identification of peptides containing the di-glycine modification (a 114.04 Da mass shift on lysine) localizes the site of ubiquitination. Linkage information can be inferred by identifying peptides derived from ubiquitin itself that contain intra-chain linkages [16].
Advantages and Limitations:
- Advantages: High-throughput and unambiguous identification of modification sites. Can reveal multiple ubiquitination sites on a single substrate.
- Limitations: Requires specialized instrumentation and expertise. The stoichiometry of ubiquitination is often low, making enrichment critical for sensitivity.
Linkage-Specific Deubiquitinase (DUB) Assays
This functional assay exploits the inherent linkage specificity of certain DUBs to probe the architecture of ubiquitin chains on your isolated substrate.
Detailed Protocol:
- Isolate Substrate: Enrich the ubiquitinated protein of interest using TUBEs.
- In Vitro DUB Treatment: Divide the purified sample into aliquots. Incubate each aliquot with a different purified, linkage-specific DUB (e.g., OTUB1 for K48-linked chains, AMSH for K63-linked chains) in an appropriate reaction buffer.
- Control Reactions: Include a control aliquot incubated with buffer alone and another with a general DUB (e.g., USP2).
- Analysis: Analyze all reactions by immunoblotting. The cleavage of ubiquitin chains by a specific DUB, observed as a collapse of the high-molecular-weight smears to the unmodified form of the protein, indicates the presence of that particular linkage type on the substrate [73].
Advantages and Limitations:
- Advantages: Provides functional, enzymatic evidence for chain linkage. Complements antibody-based approaches.
- Limitations: Requires access to purified, active DUBs. Results can be complicated if the substrate is modified by heterotypic or branched chains.
Table 2: Comparison of Key Ubiquitin Validation Techniques
| Technique | Key Application | Resolution | Throughput | Key Requirement |
|---|---|---|---|---|
| Immunoblotting | Confirm ubiquitination status and approximate chain linkage. | Low (protein-level) | Low | Specific antibodies for substrate and ubiquitin. |
| Mass Spectrometry | Identify specific modified lysine residues and chain linkages. | High (site-specific) | High | MS instrumentation and bioinformatics expertise. |
| DUB Assay | Functional validation of specific ubiquitin chain linkages. | Medium (linkage-type) | Medium | Purified, linkage-specific deubiquitinase enzymes. |
The Scientist's Toolkit: Essential Reagents for Validation
A successful validation pipeline relies on a suite of high-quality reagents and tools. The table below details essential components for the techniques described above.
Table 3: Research Reagent Solutions for Ubiquitin Validation
| Research Reagent | Function / Application | Key Characteristics |
|---|---|---|
| TUBEs (Tandem UBA Domains) | Core enrichment tool; protects and isolates polyubiquitinated proteins from native cell lysates. | High affinity for tetra-ubiquitin; protects from DUBs and proteasomal degradation [72]. |
| Linkage-Specific Ub Antibodies | Immunoblotting application to determine chain topology (e.g., K48, K63). | Specificity must be validated; can sometimes exhibit cross-reactivity [16]. |
| Anti-di-Glycine (K-ε-GG) Antibody | Mass spectrometry application for enriching ubiquitin-derived peptides to identify modification sites. | Recognizes the diglycine remnant left on modified lysine after tryptic digestion [16]. |
| Recombinant DUBs | Functional analysis of chain linkage (e.g., OTUB1 for K48, AMSH for K63). | Must be purified and enzymatically active for in vitro assays [73]. |
| Proteasome Inhibitors | Used during initial lysis to stabilize degradation-prone ubiquitinated proteins (e.g., MG132, Bortezomib). | Enhances recovery of K48-linked substrates destined for degradation. |
| DUB Inhibitors | Used during lysis to prevent deubiquitination (e.g., NEM, IAA, PR-619). | Preserves the native ubiquitination state during protein extraction [72]. |
Integrated Workflow for Ubiquitin Validation
The following diagram illustrates the integrated experimental workflow for the validation of ubiquitinated proteins, from initial isolation to final confirmation, incorporating the methodologies detailed in this note.
The study of the ubiquitin-proteasome system (UPS) is critical for understanding cellular regulation and developing therapies for diseases such as cancer and neurodegenerative disorders. Central to this research is the effective enrichment of ubiquitinated proteins from complex biological samples. Among the various tools developed for this purpose, Tandem Ubiquitin Binding Entities (TUBEs) have emerged as a powerful technology. TUBEs are engineered, high-affinity reagents composed of multiple ubiquitin-associated (UBA) domains that function as a single polypeptide to bind polyubiquitin chains with nanomolar affinity [17] [74]. Unlike single UBA domains that exhibit weak, millimolar-range affinity for ubiquitin, the tandem arrangement in TUBEs creates a synergistic, high-avidity interaction that dramatically improves enrichment efficiency [14] [74]. This article provides a comprehensive analysis of TUBE technology, detailing its optimal applications, limitations compared to alternative methodologies, and detailed protocols for implementation in modern drug discovery and basic research.
The fundamental principle behind TUBEs is their ability to recognize the three-dimensional structure of ubiquitin chains, enabling them to enrich for diverse ubiquitin chain linkages while protecting these modifications from deubiquitinating enzymes (DUBs) during cell lysis and processing [74]. This protective function is crucial for preserving the labile ubiquitin signal, which is often present at low stoichiometry within the cell. Recent advances have further refined this technology through the development of chain-specific TUBEs, which are engineered to preferentially bind particular ubiquitin linkage types, such as K48 or K63 chains, enabling researchers to dissect the specific functional consequences of different ubiquitin signals [13] [17].
Advantages of Using TUBEs
TUBEs offer several distinct advantages that make them particularly suitable for specific experimental scenarios in ubiquitin research.
Superior Affinity and Protection Against Deubiquitination
The multivalent design of TUBEs provides nanomolar affinity for polyubiquitin chains, significantly outperforming single UBA domains or traditional ubiquitin antibodies in enrichment efficiency [14] [74]. This high binding affinity is crucial for capturing low-abundance ubiquitinated species from complex lysates. Perhaps equally important is their ability to protect ubiquitin chains from deubiquitinating enzymes (DUBs) during sample preparation. The tight binding of TUBEs physically shields ubiquitin modifications from endogenous DUB activity, preserving the native ubiquitination state that might otherwise be lost during lysis and processing [74]. Experimental protocols utilizing TUBEs therefore typically include specific DUB inhibitors such as N-ethylmaleimide (NEM) at concentrations of 20 mM to ensure complete DUB inhibition and maximize ubiquitin signal preservation [74].
Capability for Linkage-Specific Analysis
A significant advancement in TUBE technology is the development of chain-specific TUBEs that can differentiate between ubiquitin linkage types. This is particularly valuable for distinguishing between degradative and non-degradative ubiquitin signaling. For example, K48-linked polyubiquitin chains typically target proteins for proteasomal degradation, while K63-linked chains are primarily involved in signaling processes related to inflammation, DNA repair, and protein trafficking [13] [17]. Research demonstrates that chain-specific TUBEs can successfully differentiate these pathways, as shown in studies of RIPK2 ubiquitination where K63-TUBEs captured L18-MDP-induced ubiquitination (inflammatory signaling), while K48-TUBEs specifically captured RIPK2 PROTAC-induced ubiquitination (degradative signaling) [13] [17]. This linkage-specific resolution provides functional insights that pan-selective enrichment methods cannot deliver.
Compatibility with Multiple Readout Platforms
TUBEs are highly versatile and compatible with various downstream analytical methods, making them suitable for multiple research applications. They can be effectively used with immunoblotting for targeted analysis of specific proteins of interest [13] [74]. When combined with mass spectrometry (TUBE-MS), they enable proteome-wide profiling of polyubiquitination changes in response to chemical treatments or physiological stimuli [74]. Additionally, their adaptability to high-throughput screening formats (such as 96-well plates) and live-cell luminescence-based assays (NanoBiT) makes them valuable for drug discovery applications where throughput and quantitative analysis are essential [13] [19]. This flexibility allows researchers to apply TUBE technology across multiple stages of investigation, from initial screening to mechanistic studies.
Table 1: Key Advantages of TUBE Technology and Their Research Implications
| Advantage | Technical Basis | Research Implication |
|---|---|---|
| High Affinity | Multivalent UBA domains with nanomolar affinity | Effective capture of low-abundance ubiquitinated targets; reduced false negatives |
| DUB Protection | Steric shielding of ubiquitin chains | Preservation of native ubiquitination status; more accurate representation of cellular state |
| Linkage Specificity | Engineered binding domains selective for specific ubiquitin linkages | Functional dissection of ubiquitin signaling (e.g., K48-degradation vs. K63-signaling) |
| Platform Flexibility | Compatible with various immobilization strategies and detection methods | Applicable from discovery proteomics to targeted validation and high-throughput screening |
Limitations and Considerations
Despite their significant advantages, TUBEs have specific limitations that researchers must consider when selecting an enrichment methodology.
Limited Effectiveness for Monoubiquitination
TUBEs are primarily optimized for capturing polyubiquitin chains and generally exhibit poorer efficiency for monoubiquitinated proteins [14]. This limitation stems from their structural design, which leverages multiple interaction surfaces that preferentially engage with the repeating units of polyubiquitin chains. Since monoubiquitination represents a substantial fraction of cellular ubiquitin modifications and regulates critical processes including endocytosis and histone function, this limitation can be significant depending on the research focus. Alternative methods such as OtUBD (a high-affinity ubiquitin-binding domain) may be more appropriate for comprehensive studies that include monoubiquitination, as OtUBD has demonstrated robust enrichment of both mono- and polyubiquitinated proteins [14].
Inability to Identify Exact Ubiquitination Sites
While TUBEs efficiently enrich ubiquitinated proteins, they do not provide direct information about specific ubiquitination sites on substrate proteins [74]. This limitation becomes important when research questions require precise mapping of modification sites, such as when studying regulatory mechanisms or designing mutants to validate functional consequences of ubiquitination. In such cases, diGly remnant enrichment methods (e.g., using antibodies against the diglycine signature left on trypsinized peptides) are more appropriate, as they specifically enable ubiquitination site identification through mass spectrometry [74]. Consequently, TUBE-based proteomics is ideal for global profiling of polyubiquitination changes, while diGly enrichment is better suited for site-specific mapping.
Potential for Co-enrichment of Interacting Proteins
Under standard (native) lysis conditions, TUBEs can co-enrich proteins that non-covalently associate with ubiquitin or ubiquitinated proteins [14]. While this can be advantageous for studying ubiquitin interactomes, it complicates the distinction between directly ubiquitinated proteins and their binding partners. This limitation can be mitigated by using semi-denaturing or fully denaturing conditions (e.g., with 4M urea or SDS) during cell lysis and washing steps, which disrupt non-covalent interactions while preserving covalent ubiquitin modifications [14] [74]. Researchers should therefore carefully select lysis conditions based on whether their goal is to study the full ubiquitin-interacting complex or specifically isolate covalently ubiquitinated proteins.
Table 2: Key Limitations of TUBEs and Strategic Workarounds
| Limitation | Impact on Research | Potential Workarounds |
|---|---|---|
| Poor Monoubiquitin Capture | Incomplete picture of ubiquitin landscape; missing monoubiquitination events | Combine with OtUBD [14] or diGly methods for comprehensive coverage |
| No Ubiquitination Site Data | Unable to determine exact modification sites on substrates | Follow-up TUBE enrichment with diGly remnant mapping for site identification [74] |
| Interacting Protein Co-enrichment | Difficulty distinguishing directly ubiquitinated proteins from binders | Use semi-denaturing/denaturing conditions (e.g., 4M urea) during lysis and washes [74] |
| Linkage Selectivity Constraints | Pan-selective TUBEs mask functional chain specificity | Employ chain-specific TUBEs when functional discrimination is required [13] |
Comparative Methodologies
To contextualize the appropriate use of TUBEs, it is essential to compare them with other established methodologies for studying protein ubiquitination.
TUBEs vs. Other Enrichment Methods
Several alternative approaches exist for enriching ubiquitinated proteins, each with distinct strengths and limitations compared to TUBEs. The OtUBD technology utilizes a high-affinity ubiquitin-binding domain from Orientia tsutsugamushi that demonstrates robust enrichment of both mono- and polyubiquitinated proteins, addressing a key limitation of TUBEs [14]. diGly remnant enrichment is particularly valuable for site-specific identification of ubiquitination but provides no information about ubiquitin chain topology [74]. Traditional immunoprecipitation with anti-ubiquitin antibodies remains widely used but often suffers from lower affinity and sensitivity compared to TUBEs [14]. Affinity-tagged ubiquitin overexpression (e.g., HA-, FLAG-, or His-tagged ubiquitin) enables efficient purification but creates non-physiological conditions that may produce artifactual results [14].
Table 3: Comparison of Key Methodologies for Ubiquitin Enrichment
| Method | Key Advantage | Primary Limitation | Ideal Application |
|---|---|---|---|
| TUBEs | High-affinity polyubiquitin capture; DUB protection; linkage-specific options | Poor monoubiquitin capture; no site information | Polyubiquitination profiling; functional studies of chain types; protective enrichment |
| OtUBD | Effective mono- and polyubiquitin enrichment | Less established for chain-specific work | Comprehensive ubiquitome studies including monoubiquitination |
| diGly Remnant Enrichment | Precise ubiquitination site mapping | No chain topology information; destroys protein context | Site-specific ubiquitination analysis and mapping |
| Tagged Ubiquitin Overexpression | High sensitivity and yield | Non-physiological artifacts; overexpression effects | Controlled systems where endogenous detection fails |
| Traditional Immunoprecipitation | Widely accessible; works with endogenous ubiquitin | Lower affinity/sensitivity; limited DUB protection | Initial screening when specialized reagents unavailable |
Guidelines for Method Selection Based on Research Goals
Selecting the appropriate ubiquitin enrichment method depends primarily on the specific research question and experimental context. For studies focused specifically on polyubiquitination, particularly when investigating chain-type specificity or needing protection from DUBs, TUBEs are the superior choice [13] [74]. For comprehensive ubiquitome profiling that includes monoubiquitination, OtUBD or diGly methods may be more appropriate [14]. When precise ubiquitination site mapping is the primary goal, diGly remnant enrichment is indispensable [74]. For high-throughput screening of compound libraries targeting the UPS, TUBE-based assays in microtiter plate formats provide an optimal combination of throughput and functional information [13] [19]. In validation studies following proteomic discovery, a combination of methods is often ideal—using TUBE-based immunoblotting to confirm polyubiquitination status followed by diGly mapping for site resolution.
Experimental Protocols and Applications
Detailed Protocol: TUBE-Based Enrichment for Immunoblotting
This protocol describes the enrichment of ubiquitinated proteins from cell lysates using TUBE-conjugated magnetic beads, optimized for subsequent detection by immunoblotting [13] [74].
Reagents and Solutions:
- Lysis Buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 1 mM EDTA, 20 mM N-ethylmaleimide (NEM), 10 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), and complete EDTA-free protease inhibitor cocktail [74]
- Wash Buffer: Lysis buffer supplemented with 4 M urea [74]
- TUBE-Conjugated Magnetic Beads: Pan-selective or chain-specific TUBEs immobilized on streptavidin-coated magnetic beads [13] [74]
- Elution Buffer: 1× SDS-PAGE sample buffer containing 50 mM DTT
Procedure:
- Cell Lysis: Harvest cells and lyse in ice-cold lysis buffer (1 mL per 10⁷ cells). Incubate on ice for 15 minutes with occasional vortexing.
- Clarification: Centrifuge lysates at 16,000 × g for 15 minutes at 4°C. Transfer supernatant to a new tube.
- Protein Quantification: Determine protein concentration using Bradford or BCA assay.
- Enrichment: Incubate 500-1000 μg of protein lysate with 50 μL of TUBE-conjugated magnetic beads for 2 hours at 4°C with end-over-end mixing.
- Washing: Pellet beads and wash three times with 1 mL of wash buffer (4 M urea), followed by one wash with standard lysis buffer.
- Elution: Elute bound proteins by adding 50 μL of Elution Buffer and heating at 95°C for 10 minutes.
- Analysis: Resolve eluates by SDS-PAGE and proceed with immunoblotting using target-specific antibodies.
Detailed Protocol: TUBE-MS for Proteomic Profiling
This protocol describes TUBE enrichment coupled with mass spectrometry (TUBE-MS) for proteome-wide analysis of polyubiquitination changes [74].
Reagents and Solutions:
- Semi-denaturing Lysis Buffer: 50 mM Tris-HCl (pH 7.5), 1% SDS, 150 mM NaCl, 20 mM NEM, 10 mM TCEP, with protease inhibitors
- Urea Wash Buffer: 50 mM Tris-HCl (pH 7.5), 4 M urea, 150 mM NaCl
- ABC Buffer: 50 mM ammonium bicarbonate, pH 8.0
- Trypsin/Lys-C mix for protein digestion
Procedure:
- Cell Lysis: Lyse cells in semi-denaturing lysis buffer with heating at 95°C for 10 minutes to dissociate non-covalent interactions.
- Digestion and Dilution: Dilute lysate 1:10 with 50 mM Tris-HCl (pH 7.5) to reduce SDS concentration to 0.1%.
- TUBE Enrichment: Incubate diluted lysate (equivalent to 1-2 mg protein) with biotinylated TUBEs pre-bound to streptavidin magnetic beads for 2 hours at 4°C.
- Stringent Washing: Wash beads sequentially with:
- Urea Wash Buffer (twice)
- Standard Lysis Buffer (once)
- 50 mM ammonium bicarbonate buffer (twice)
- On-Bead Digestion: Resuspend beads in ABC buffer and digest proteins with Trypsin/Lys-C mix overnight at 37°C.
- Peptide Collection: Collect supernatant containing digested peptides and acidify with formic acid.
- LC-MS/MS Analysis: Analyze peptides by liquid chromatography coupled to tandem mass spectrometry.
Application Example: Studying PROTAC-Induced Ubiquitination
PROTACs (Proteolysis Targeting Chimeras) are heterobifunctional molecules that recruit E3 ligases to target proteins, inducing their ubiquitination and degradation. TUBE-based assays are particularly valuable for characterizing PROTAC mechanism of action [13] [74].
Experimental Design:
- Treat cells with PROTAC of interest (e.g., MZ1 for BRD2 degradation) and appropriate control compounds.
- Co-treat with proteasome inhibitor (e.g., Carfilzomib, 1 μM for 1 hour) before harvesting to stabilize ubiquitinated proteins.
- Prepare lysates using protective lysis buffer with 20 mM NEM.
- Enrich ubiquitinated proteins using pan-selective TUBEs to capture overall ubiquitination or K48-specific TUBEs to specifically monitor degradative ubiquitination.
- Analyze by immunoblotting with antibodies against the target protein to detect higher molecular weight smears characteristic of polyubiquitination.
This approach enables direct confirmation of PROTAC-induced ubiquitination and can differentiate between productive K48-linked ubiquitination versus non-degradative ubiquitin chain formation.
Successful implementation of TUBE-based methodologies requires specific reagents and tools optimized for ubiquitin research.
Table 4: Essential Research Reagents for TUBE-Based Ubiquitin Enrichment
| Reagent/Tool | Function | Application Notes |
|---|---|---|
| TUBE Reagents | High-affinity enrichment of polyubiquitin chains | Available as pan-selective or linkage-specific (K48, K63) formats; can be biotinylated for bead immobilization [13] [74] |
| DUB Inhibitors | Preserve ubiquitin signals during processing | N-ethylmaleimide (NEM; 20 mM) effectively inhibits cysteine-based DUBs during lysis [74] |
| Semi-denaturing Buffers | Disrupt non-covalent interactions while preserving ubiquitination | 4 M urea in wash buffers reduces co-enrichment of ubiquitin-binding proteins [74] |
| Chain-specific TUBEs | Differentiation of ubiquitin linkage types | K48-TUBEs for degradative ubiquitination; K63-TUBEs for signaling ubiquitination [13] [17] |
| TUBE-Coated Plates | High-throughput screening format | 96-well microplates pre-coated with TUBEs enable automated processing and screening [13] |
| Compatibile Lysis Buffers | Maintain protein integrity while inhibiting DUBs | Should include NEM (20 mM) and protease inhibitors; may include SDS for semi-denaturing conditions [74] |
Visualizing Experimental Workflows
The following diagrams illustrate key experimental setups and methodological relationships in TUBE-based ubiquitin research.
Diagram 1: TUBE Experimental Workflow Selection. This flowchart guides researchers through key decision points in designing TUBE-based experiments, including lysis conditions and choice of detection method based on research objectives [13] [14] [74].
Diagram 2: Method Selection Decision Tree. This flowchart provides a systematic approach for selecting the most appropriate ubiquitin enrichment methodology based on specific research requirements and questions [13] [14] [74].
TUBE technology represents a powerful and versatile approach for studying protein ubiquitination, particularly when research focuses on polyubiquitin chain biology, requires protection of labile ubiquitin modifications, or demands differentiation between ubiquitin linkage types with specific functional consequences. Its strengths in high-affinity enrichment, DUB protection, and compatibility with various detection platforms make it especially valuable for drug discovery applications targeting the ubiquitin-proteasome system, such as characterizing PROTAC-mediated ubiquitination. However, researchers must also recognize its limitations regarding monoubiquitination capture and site identification, where alternative methods like OtUBD or diGly remnant enrichment may be more appropriate. The optimal application of TUBEs therefore depends on carefully matching methodological capabilities to specific research questions, and in many cases, combining TUBE-based approaches with complementary methodologies provides the most comprehensive insight into the complex landscape of protein ubiquitination.
Conclusion
Tandem Ubiquitin-Binding Entities represent a paradigm shift in ubiquitin research, moving the field beyond the limitations of traditional methods. By providing a robust, high-affinity means to isolate, enrich, and—crucially—protect polyubiquitinated proteins under native conditions, TUBEs have unlocked new avenues for discovery. The key takeaways are their unparalleled ability to shield substrates from deubiquitinating enzymes and the proteasome, their flexibility in application from basic research to proteomic screens, and their growing specificity for distinct polyubiquitin linkages. The future of TUBE technology is bright, with implications for accelerating drug discovery, particularly in targeting the ubiquitin system for therapies in cancer and neurodegeneration. As the toolbox expands with new linkage-specific TUBEs and related entities like SUBEs for SUMOylation research, these molecular traps will remain indispensable for unraveling the complexities of the ubiquitin code and translating these findings into clinical advancements.
References
- 1. The Ubiquitin Tale: Current Strategies and Future Challenges [https://pmc.ncbi.nlm.nih.gov/articles/PMC11406696/]
- 2. The ubiquitin system: from cell signalling to disease ... [https://www.nature.com/articles/s41418-020-00703-w]
- 3. Ubiquitin modifications | Cell Research [https://www.nature.com/articles/cr201639]
- 4. Current methodologies in protein ubiquitination ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9373315/]
- 5. Tandem Ubiquitin Binding Entities (TUBEs) as Tools to ... [https://pubmed.ncbi.nlm.nih.gov/34432245/]
- 6. Tandem Ubiquitin Binding Entities (TUBEs) [https://lifesensors.com/our-technologies/tandem-ubiquitin-binding-entities-tubes/]
- 7. Post-translational regulation of ubiquitin signaling - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6548142/]
- 8. An inventory of crosstalk between ubiquitination and other ... [https://www.sciencedirect.com/science/article/pii/S258900422300353X]
- 9. What Is Used to Enrich Ubiquitin Proteins? [https://www.mtoz-biolabs.com/what-is-used-to-enrich-ubiquitin-proteins.html]
- 10. Ubiquitin binding domains — from structures to functions - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7359374/]
- 11. Ubiquitin-binding domains - PMC - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC1615911/]
- 12. Enhanced Purification of Ubiquitinated Proteins by ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4824862/]
- 13. Unraveling chain specific ubiquitination in cells using ... [https://www.nature.com/articles/s41598-025-07242-9]
- 14. Use of a High-Affinity Ubiquitin-Binding Domain to Detect and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12423275/]
- 15. A High-Throughput Method for Specific, Rapid, Precise and ... [https://www.sciencedirect.com/science/article/abs/pii/S0039914025015681]
- 16. Current methodologies in protein ubiquitination characterization [https://cellandbioscience.biomedcentral.com/articles/10.1186/s13578-022-00870-y]
- 17. Unraveling Chain specific Ubiquitination in Cells Using ... [https://lifesensors.com/unraveling-chain-specific-ubiquitination-in-cells-using-tandem-ubiquitin-binding-entities-in-a-high-throughput-assay/]
- 18. Integrative analysis of the ubiquitin proteome isolated ... [https://www.sciencedirect.com/science/article/abs/pii/S1874391911006683]
- 19. A Novel Luminescence-Based High-Throughput Approach ... [https://www.sciencedirect.com/science/article/pii/S2472555222065571]
- 20. TUBE (Tandem Ubiquitin Binding Entity) (LifeSensors) [https://www.nacalai.com/global/reagent/distributed/12.html]
- 21. Polyubiquitinated Proteins Isolation- M1 TUBE [https://lifesensors.com/product/polyubiquitinated-proteins-isolation-m1-tube/]
- 22. UM402M: TUBE 2 (Magnetic Beads) [https://lifesensors.com/product/um402m-tube-2-magnetic-beads/]
- 23. K48-TUBEs [https://lifesensors.com/a-new-generation-of-tubes/]
- 24. Polyubiquitinated Proteins Enrichment ( K HF) - LifeSensors 48 TUBE [https://lifesensors.com/product/polyubiquitinated-proteins-enrichment-k48-tube-hf/]
- 25. Unraveling chain specific ubiquitination in cells using tandem ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12217038/]
- 26. - and K -linked ubiquitin chain interactome reveals branch- and... 48 K 63 [https://pmc.ncbi.nlm.nih.gov/articles/PMC11109483/]
- 27. The Molecular Toolbox for Linkage Type‐Specific Analysis ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12118340/]
- 28. Integrative analysis of the ubiquitin proteome isolated ... [https://pubmed.ncbi.nlm.nih.gov/22178446/]
- 29. Optimising methods for the preservation, capture and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4709362/]
- 30. Protocol of Preparation of Ubiquitinated Protein Samples [https://www.creativebiomart.net/resource/principle-protocol-protocol-of-preparation-of-ubiquitinated-protein-samples-428.htm]
- 31. Large-Scale Identification of Ubiquitination Sites by Mass ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4725055/]
- 32. How to Better Study Ubiquitination [https://www.ptglab.com/news/blog/how-to-better-study-ubiquitination/?srsltid=AfmBOop4zLE4qIsX-XuX3h7QD8kavW3XrzA-eM5cx4X36aj0APO4SIch]
- 33. Comprehensive Guide to Cell Lysis and Protein Extraction ... [https://www.creative-proteomics.com/resource/comprehensive-guide-cell-lysis-protein-extraction.htm]
- 34. LifeSensors Targeted Protein Degredation [https://www.2bscientific.com/blog/2025/november/lifesensors-targeted-protein-degredation]
- 35. Immunoprecipitation and co-immunoprecipitation protocol [https://www.abcam.com/en-us/technical-resources/protocols/immunoprecipitation]
- 36. Fundamentals and Visualization of Enrichment Analysis in ... [https://www.protocols.io/view/enrichment-analysis-fundamentals-and-visualization-c5rxy57n.html]
- 37. Unveiling the Power of TUBEs: Revolutionizing Ubiquitin ... [https://lifesensors.com/unveiling-the-power-of-tubes-revolutionizing-ubiquitin-research/]
- 38. Mass Spectrometry Proteomics using TUBEs [https://lifesensors.com/mass-spectromtry-proteomics-using-tubes/]
- 39. BioE3 identifies specific substrates of ubiquitin E3 ligases [https://www.nature.com/articles/s41467-023-43326-8]
- 40. Ubiquitination and deubiquitination in cancer [https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-024-02046-3]
- 41. The multifaceted role of PUMA in cell death pathways [https://biosignaling.biomedcentral.com/articles/10.1186/s12964-025-02112-3]
- 42. Ubiquitination of transcription factors in cancer [https://pmc.ncbi.nlm.nih.gov/articles/PMC12330937/]
- 43. Decoding the ubiquitin network: molecular mechanisms and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12492556/]
- 44. Recent advances in small molecule inhibitors of ... [https://www.sciencedirect.com/science/article/abs/pii/S0223523425000893]
- 45. Small Molecule-Induced Alterations of Protein ... [https://pubmed.ncbi.nlm.nih.gov/40444580/]
- 46. Pre-analytical drivers of bias in bead-enriched plasma ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12603263/]
- 47. Theoretical Investigation of the Material Usage During On- ... [https://www.mdpi.com/1420-3049/30/15/3245]
- 48. Optimized Automated Workflow for BioID Improves ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11460324/]
- 49. Protocol for tandem enrichment of ubiquitinated, ... [https://www.sciencedirect.com/science/article/pii/S2666166725003508]
- 50. A comprehensive method for detecting ubiquitinated ... [https://pubmed.ncbi.nlm.nih.gov/25827227/]
- 51. A substrate-trapping strategy to find E3 ubiquitin ligase ... [https://www.nature.com/articles/s42003-020-01328-y]
- 52. Designing Effective Data Visualizations [https://guides.library.jhu.edu/datavisualization/design]
- 53. Best Practices - Data Visualization [https://libguides.gwu.edu/dataviz/best_practices]
- 54. Text has enhanced contrast | ACT Rule | WAI [https://www.w3.org/WAI/standards-guidelines/act/rules/09o5cg/]
- 55. Color Contrast | Web Accessibility Checklist [https://dequeuniversity.com/checklists/web/color-contrast]
- 56. Proper Reagent Storage and Handling | Updated 2025 [https://www.stressmarq.com/blog/proper-reagent-storage-and-handling/?srsltid=AfmBOoqtU0RlwZNsm-e1rwQ8fL3M6FxR5GhHIW1BZPoXdUL_wS5GR_uk]
- 57. Safe Lab Reagent Storage Guide | Best Practices 2025 [https://www.labdisposable.com/best-practices-for-storing-lab-reagents-safe-compliant-and-organized-laboratory-storage/]
- 58. Best Practices for Proper Storage and Handling of ... [https://www.needle.tube/resources-20/Best-Practices-for-Proper-Storage-and-Handling-of-Reagents-and-Supplies-in-a-Clinical-Chemistry-Lab-in-the-United-States]
- 59. Ubiquitin diGLY proteomics as an approach to identify and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6791129/]
- 60. Data-independent acquisition method for ubiquitinome ... [https://www.nature.com/articles/s41467-020-20509-1]
- 61. Proteomic approaches to study ubiquitinomics [https://www.sciencedirect.com/science/article/abs/pii/S1874939923000354]
- 62. Evaluation of Enrichment Techniques for Mass Spectrometry [https://pmc.ncbi.nlm.nih.gov/articles/PMC1867451/]
- 63. Improvement of ubiquitylation site detection by Orbitrap ... [https://www.sciencedirect.com/science/article/pii/S1874391917303718]
- 64. TUBE 1 (Tandem Ubiquitin Binding Entity) [https://lifesensors.com/product/um101-tube-1-tandem-ubiquitin-binding-entity-lifesensors-product/]
- 65. TUBE-based Mass Spec Proteomics [https://lifesensors.com/tube-based-mass-spectrometry/]
- 66. TUBEs for Detection [https://lifesensors.com/product-category/tubes-ubiquitin-affinity-matrix/tubes-for-detection/]
- 67. 12-Tube Magnetic Separation Rack #14654 [https://www.cellsignal.com/products/chip-kits-reagents/12-tube-magnetic-separation-rack/14654?srsltid=AfmBOorYTDecvAPoXS_fbWR_Yep3qv5irReDMA4qMH_BhCEHfNEzUJwQ]
- 68. 6-Tube Magnetic Separation Rack #7017 [https://www.cellsignal.com/products/chip-kits-reagents/6-tube-magnetic-separation-rack/7017?srsltid=AfmBOorU6AWl8iQcmGrmpMYTHx9fPNlL6d_nHfyxAjM-gUMOKeSk7uZI]
- 69. Recent Advances on Cell Culture Platforms for In Vitro Drug ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11468737/]
- 70. Platform Technology Designation Program for Drug ... [https://www.fda.gov/regulatory-information/search-fda-guidance-documents/platform-technology-designation-program-drug-development]
- 71. Ubiquitination detection techniques - PMC - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC10625345/]
- 72. Efficient protection and isolation of ubiquitylated proteins ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2775171/]
- 73. Ubiquitin Linkage Specificity of Deubiquitinases ... [https://pubmed.ncbi.nlm.nih.gov/32240951/]
- 74. Small Molecule‐Induced Alterations of Protein ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12322627/]