UBD vs. Antibody-Based Methods: A Comprehensive Guide to Evaluating Enrichment Efficiency for Protein Ubiquitination
This article provides a systematic evaluation of Ubiquitin-Binding Domain (UBD)-based and antibody-based methods for enriching ubiquitinated proteins, a critical step in proteomics and drug discovery.
UBD vs. Antibody-Based Methods: A Comprehensive Guide to Evaluating Enrichment Efficiency for Protein Ubiquitination
Abstract
This article provides a systematic evaluation of Ubiquitin-Binding Domain (UBD)-based and antibody-based methods for enriching ubiquitinated proteins, a critical step in proteomics and drug discovery. Tailored for researchers and drug development professionals, we compare the foundational principles, methodological workflows, and key performance metrics of these techniques. The content delves into common optimization challenges, offers practical troubleshooting advice, and presents a framework for the rigorous, comparative validation of enrichment efficiency. By synthesizing current research and technological advancements, this guide aims to equip scientists with the knowledge to select and optimize the most appropriate method for their specific research goals in biomedicine and therapeutics.
The Building Blocks of Ubiquitin Enrichment: Understanding UBDs and Antibodies
Protein ubiquitination, a fundamental post-translational modification, regulates virtually all cellular processes in eukaryotic cells. This enzymatic process involves the covalent attachment of ubiquitin to target proteins, modulating their stability, activity, localization, and interactions [1]. The versatility of ubiquitination stems from its ability to form diverse chain topologies, including homotypic, mixed, and branched polymers with distinct biological functions [2]. Understanding ubiquitination is crucial for elucidating numerous disease mechanisms, as dysregulation of ubiquitin signaling is implicated in cancer, neurodegenerative disorders, and inflammatory conditions [1]. This review systematically compares contemporary methodologies for studying ubiquitination, with particular emphasis on evaluating the enrichment efficiency of ubiquitin-binding domain (UBD)-based versus antibody-based approaches, providing researchers with critical insights for experimental design.
The Ubiquitination Machinery and Signaling Complexity
Ubiquitination involves a sophisticated enzymatic cascade comprising E1 (activating), E2 (conjugating), and E3 (ligating) enzymes that collectively coordinate the covalent attachment of the 76-amino acid ubiquitin protein to substrate lysine residues [3]. The human genome encodes approximately 2 E1 enzymes, 40 E2 enzymes, and over 600 E3 ligases, which confer substrate specificity [4]. This process is reversible through the action of deubiquitinating enzymes (DUBs), with approximately 100 encoded in the human genome, maintaining ubiquitination homeostasis [4].
The complexity of ubiquitin signaling arises from the ability of ubiquitin itself to be modified on its N-terminal methionine (M1) and seven lysine residues (K6, K11, K27, K29, K33, K48, K63), generating diverse chain architectures with distinct functional consequences [3] [2]. K48-linked chains primarily target substrates for proteasomal degradation, while K63-linked chains and M1-linear chains regulate non-degradative functions including signal transduction, DNA repair, and selective autophagy [4] [3]. More recently, branched ubiquitin chains containing multiple linkage types have emerged as specialized signals that expand the ubiquitin code's functional repertoire [2].
The biological outcomes of ubiquitination are determined through recognition by proteins containing ubiquitin-binding domains (UBDs) that decipher the chain topology and transmit appropriate downstream signals [3]. This sophisticated system precisely controls protein homeostasis, DNA repair, cell cycle progression, immune responses, and numerous other fundamental cellular processes [1].
Methodological Approaches for Ubiquitination Analysis
Studying ubiquitination presents significant technical challenges due to the low stoichiometry of modified proteins, the diversity of modification sites, and the structural complexity of ubiquitin chains. This section compares the principal methodologies for ubiquitin enrichment, focusing on UBD-based versus antibody-based approaches.
Antibody-Based Enrichment Methods
Antibody-based approaches utilize ubiquitin-specific antibodies to isolate ubiquitinated proteins or peptides from complex biological samples. The most widely employed antibodies include:
- Pan-specific antibodies (P4D1, FK1/FK2) that recognize all ubiquitin linkages [4]
- Linkage-specific antibodies that selectively bind particular chain types (M1-, K11-, K27-, K48-, K63-linkages) [4]
- diGly remnant antibodies that target the characteristic diglycine signature left on trypsinized peptides after ubiquitination [5]
The diGly antibody-based approach has been particularly powerful when combined with advanced mass spectrometry techniques. A recent optimized workflow combining diGly antibody enrichment with data-independent acquisition (DIA) mass spectrometry identified approximately 35,000 distinct diGly peptides in single measurements of proteasome inhibitor-treated cells, doubling the identification capacity of previous data-dependent acquisition methods [5]. This approach demonstrated exceptional quantitative accuracy, with 45% of diGly peptides showing coefficients of variation (CVs) below 20% across replicates [5].
Ubiquitin-Binding Domain (UBD)-Based Enrichment Methods
UBD-based methodologies exploit natural ubiquitin receptors to capture ubiquitinated proteins. These include:
- Tandem ubiquitin-binding entities (TUBEs) that combine multiple UBDs to enhance affinity [4]
- UBD-fusion proteins that utilize specific ubiquitin-recognition domains from proteins such as E3 ligases, DUBs, and ubiquitin receptors [4]
- Modified UBD constructs engineered for improved specificity toward particular chain topologies [3]
Single UBDs typically exhibit low affinity for ubiquitin, limiting their utility for efficient enrichment. However, tandem-repeated UBD designs significantly enhance binding capacity, making them valuable tools for capturing endogenous ubiquitination without genetic manipulation [4]. UBD-based approaches are particularly advantageous for preserving labile ubiquitin signals during extraction, as many TUBEs offer protection against deubiquitinating enzyme activity [4].
Comparative Efficiency of Enrichment Methods
Table 1: Comparative Analysis of Ubiquitin Enrichment Methodologies
| Method Feature | Antibody-Based | UBD-Based | Ub-Tagging |
|---|---|---|---|
| Throughput | High-throughput [5] | Moderate | Low to moderate [4] |
| Specificity | High (especially linkage-specific antibodies) [4] | Variable depending on UBD | High for tagged systems [4] |
| Endogenous Application | Yes (directly applicable) [4] | Yes (directly applicable) [4] | Requires genetic manipulation [4] |
| Linkage Resolution | Excellent with linkage-specific antibodies [4] | Moderate to good | Limited without additional strategies |
| Typical Identification Yield | ~35,000 diGly peptides (single shot) [5] | Not explicitly quantified in results | ~750 ubiquitination sites (Strep-tag) [4] |
| Key Advantages | High sensitivity and specificity; commercial availability | Preservation of labile modifications; recognition of specific architectures | No antibody cross-reactivity issues |
| Main Limitations | High cost; potential non-specific binding | Lower affinity for some constructs; less established | Cannot use in clinical samples; artificial system [4] |
Table 2: Quantitative Performance Comparison of diGly Enrichment Methods
| Performance Metric | DDA with diGly Antibodies | DIA with diGly Antibodies | Ub Tagging (Strep/His) |
|---|---|---|---|
| Identifications (single shot) | ~20,000 diGly peptides [5] | ~35,000 diGly peptides [5] | ~750 ubiquitination sites [4] |
| Quantitative Precision (CV <20%) | 15% of peptides [5] | 45% of peptides [5] | Not specified |
| Reproducibility | Moderate | High | Moderate |
| Sample Requirement | 1mg peptide input [5] | 1mg peptide input [5] | Requires engineered cells [4] |
| Applicability to Tissues | Directly applicable [4] | Directly applicable [4] | Not applicable [4] |
Experimental Protocols for Ubiquitination Analysis
Protocol: diGly Antibody-Based Ubiquitinome Profiling
This protocol outlines the optimized workflow for large-scale ubiquitin site identification using diGly antibody enrichment coupled with DIA mass spectrometry [5].
Sample Preparation:
- Harvest cells or tissues and lyse in denaturing buffer (e.g., 8M urea, 50mM Tris-HCl pH 8.0) with protease inhibitors and DUB inhibitors to preserve ubiquitin signals.
- Reduce disulfide bonds with 5mM dithiothreitol (60°C, 30min) and alkylate with 10mM iodoacetamide (room temperature, 30min in darkness).
Protein Digestion:
- Dilute urea concentration to 2M and digest proteins with Lys-C (1:100 enzyme:protein, 4h, room temperature).
- Further dilute to 1M urea and digest with trypsin (1:100 enzyme:protein, overnight, 37°C).
Peptide Desalting:
- Acidify peptides with trifluoroacetic acid (TFA) to pH <3.
- Desalt using C18 solid-phase extraction cartridges or StageTips.
diGly Peptide Enrichment:
- Resuspend peptides in immunoaffinity purification (IAP) buffer (1mg peptide input recommended).
- Incubate with anti-diGly antibody (31.25μg recommended) conjugated to protein A/G beads for 2h at 4°C with gentle agitation.
- Wash beads extensively with IAP buffer followed by water.
Peptide Elution:
- Elute diGly peptides with 0.1% TFA (2 × 100μL).
- Concentrate and desalt eluted peptides using C18 StageTips.
Mass Spectrometry Analysis:
- Analyze using optimized DIA method with 46 precursor isolation windows covering 400-1000 m/z range.
- Use MS2 resolution of 30,000 for improved identification.
- Employ comprehensive spectral libraries (≥90,000 diGly peptides) for optimal data extraction [5].
Protocol: UBD-Based Enrichment of Ubiquitinated Proteins
This protocol describes the isolation of ubiquitinated proteins using tandem ubiquitin-binding entities (TUBEs) [4].
Cell Lysis:
- Lyse cells in NP-40 or RIPA buffer containing complete protease inhibitors and 10-20mM N-ethylmaleimide (DUB inhibitor).
- Centrifuge at 20,000 × g for 15min to remove insoluble material.
TUBE Affinity Purification:
- Incubate cleared lysate with agarose- or magnetic bead-conjugated TUBEs (2-4μg TUBE per mg total protein) for 4h at 4°C with gentle rotation.
- Wash beads 3-4 times with lysis buffer containing 300-500mM NaCl to reduce non-specific binding.
Elution Options:
- For downstream immunoblotting: Elute with 2× Laemmli buffer at 95°C for 10min.
- For proteomics: Elute with acidic conditions (0.1M glycine pH 2.5) or competitive elution with free ubiquitin (1mg/mL).
Analysis:
- Process eluates for immunoblotting with ubiquitin-specific antibodies.
- For mass spectrometry, proceed with standard protein digestion and peptide cleanup.
Visualization of Ubiquitination Pathways and Methodologies
Diagram 1: Ubiquitin Signaling Cascade. The sequential action of E1, E2, and E3 enzymes mediates the attachment of ubiquitin to protein substrates, forming monoubiquitination, polyubiquitination, or branched chains with distinct functional consequences [3] [2].
Diagram 2: Ubiquitin Enrichment Method Comparison. Three principal methodologies for isolating ubiquitinated proteins or peptides, each with distinct advantages, limitations, and typical identification yields based on current methodologies [4] [5].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Essential Research Reagents for Ubiquitination Studies
| Reagent Category | Specific Examples | Primary Applications | Key Features |
|---|---|---|---|
| Ubiquitin Antibodies | P4D1, FK1/FK2 (pan-specific); K48-/K63-linkage specific [4] | Immunoblotting, immunofluorescence, immunoprecipitation | Recognition of all ubiquitinated proteins or specific chain types |
| diGly Antibodies | PTMScan Ubiquitin Remnant Motif Kit [5] | Mass spectrometry-based ubiquitinome profiling | Enrichment of tryptic peptides containing K-ε-GG remnant |
| TUBEs (Tandem Ubiquitin-Binding Entities) | Recombinant proteins with multiple UBDs [4] | Purification of endogenous ubiquitinated proteins | Protection from DUB activity; recognition of various chain types |
| Ubiquitin Variants | Wild-type Ub, K-only mutants, M1-only mutant [3] | Mechanism studies, linkage specificity assays | Defined chain formation in biochemical assays |
| DUB Inhibitors | PR-619, N-ethylmaleimide [4] | Preservation of ubiquitin signals during extraction | Broad-spectrum or specific DUB inhibition |
| Activity-Based Probes | Ubiquitin-dehydroalanine (Ub-Dha) [3] | DUB activity profiling, identification | Covalent trapping of active DUBs |
| Epitope-Tagged Ubiquitin | His-Ub, HA-Ub, Strep-Ub, GFP-Ub [4] [3] | Affinity purification, live-cell imaging | Enrichment of ubiquitinated proteins from engineered cells |
The comprehensive analysis of protein ubiquitination remains challenging yet essential for understanding its critical roles in cellular regulation and disease. Method selection should be guided by specific research questions, with antibody-based approaches offering superior sensitivity and specificity for site identification, while UBD-based methods provide advantages for preserving labile modifications and studying specific chain architectures. The ongoing development of improved mass spectrometry acquisition strategies like DIA has dramatically enhanced the depth and quantitative accuracy of ubiquitinome analyses [5]. Future methodological advances will likely focus on improving linkage-specific enrichment, resolving branched chain complexity, and enabling spatial analysis of ubiquitination through advanced microscopy techniques [3] [2]. As these methodologies continue to evolve, they will undoubtedly uncover new dimensions of ubiquitin signaling and its therapeutic potential in human disease.
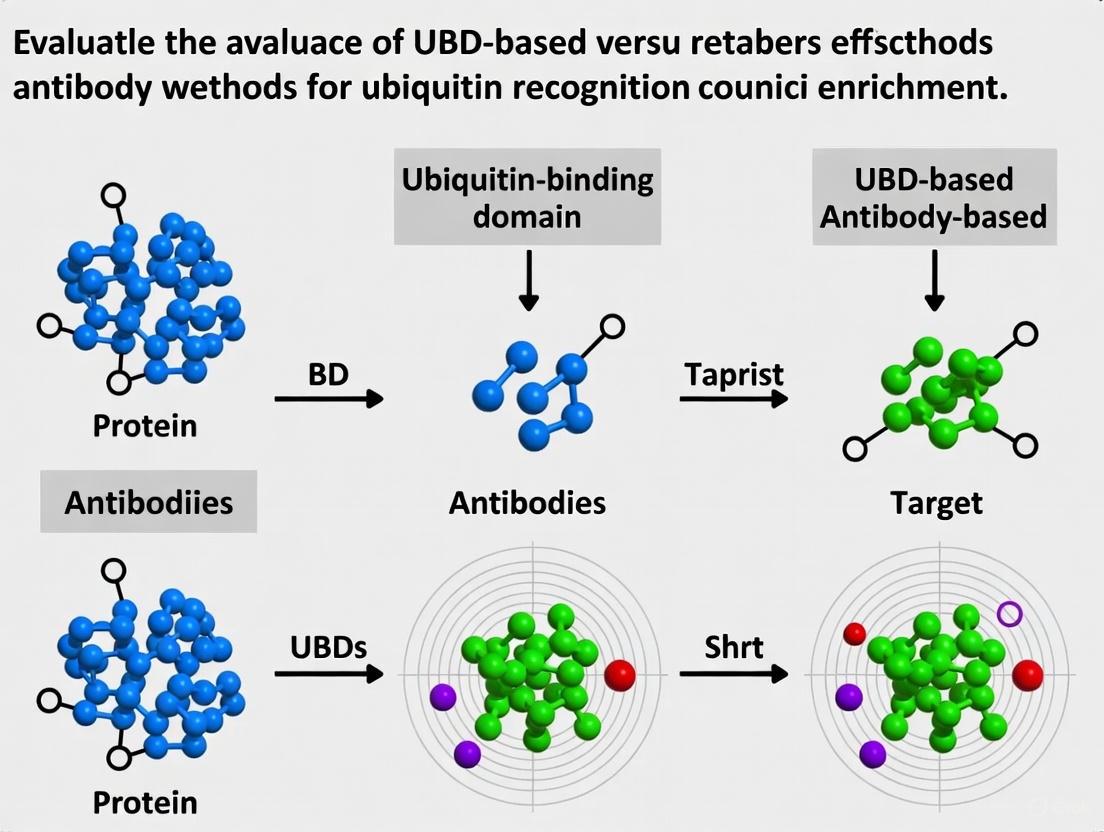
In the study of proteins and their post-translational modifications, the targeted enrichment of specific molecules from complex biological mixtures is a fundamental prerequisite for detailed analysis. This is particularly true for low-abundance targets, where enrichment efficiency directly determines the sensitivity, specificity, and ultimate success of downstream analytical methods like mass spectrometry. Two principal strategies have emerged for this purpose: antibody-based enrichment and ubiquitin-binding domain (UBD)-based enrichment. Antibody-based methods leverage the specific recognition of epitopes—distinct molecular surfaces on the target—by immunoglobulin-based reagents. UBD-based methods utilize naturally occurring protein domains that have evolved to bind ubiquitin or ubiquitinated substrates with high affinity. The core principles governing the efficiency of these techniques are epitope recognition, binding affinity, and linkage bias. Understanding the interplay of these principles is essential for selecting the appropriate method for a given research application, particularly in the growing field of ubiquitin research where both approaches are widely used. This guide provides an objective comparison of these methodologies, supported by experimental data and detailed protocols.
Core Principles of Enrichment Technologies
Principle 1: Epitope Recognition and Specificity
Epitope recognition refers to the specific interaction between an affinity reagent (e.g., an antibody) and a distinct region on its target molecule. The nature and accessibility of this epitope are primary determinants of enrichment success.
- Antibody-Based Recognition: Traditional antibodies typically recognize a specific, three-dimensional epitope on a target protein or modification. For ubiquitin research, some antibodies (e.g., P4D1, FK1/FK2) are designed to recognize all ubiquitinated substrates, while others are linkage-specific, targeting unique structural features of particular polyubiquitin chain types (e.g., K48-, K63-, or M1-linked chains) [4]. A key limitation is that modifications near the antibody's epitope, such as the conjugation of small molecule drugs in Antibody-Drug Conjugates (ADCs), can cause steric hindrance and significantly reduce binding efficiency [6]. Innovations like Multiepitope Recognition Technology (MERT) aim to overcome this by simultaneously targeting multiple distinct regions of a protein, such as both the complementarity-determining regions (CDRs) and non-CDR areas of an antibody. This approach has been shown to improve specificity and binding capacity by ten to a hundred-fold compared to mono-epitope methods [6].
- UBD-Based Recognition: UBDs, such as the recently characterized OtUBD from Orientia tsutsugamushi, recognize the conserved ubiquitin fold itself [7]. Their specificity is not for a linear sequence but for the three-dimensional surface of ubiquitin. This allows UBDs to bind a wide range of ubiquitylated proteins, provided the ubiquitin moiety is accessible. Tandem-repeated UBDs (TUBEs) increase the avidity of this interaction by engaging multiple ubiquitin molecules within a polyubiquitin chain simultaneously [4] [7].
Principle 2: Binding Affinity and Avidity
Affinity measures the strength of a single interaction between a binding site and its ligand, while avidity refers to the combined strength of multiple simultaneous interactions.
- Intrinsic Affinity: Individual UBDs typically bind ubiquitin with relatively low, micromolar-range affinity (Kd) [7]. In contrast, high-quality monoclonal antibodies can achieve nanomolar or even picomolar affinity for their targets, making them individually stronger binders.
- The Avidity Advantage: To compensate for modest intrinsic affinity, UBDs are often deployed as tandem ubiquitin-binding entities (TUBEs), where multiple UBDs are fused in a single polypeptide. This configuration creates a powerful avidity effect, dramatically increasing the apparent affinity for polyubiquitinated substrates. This makes TUBEs exceptionally effective for enriching proteins modified with longer ubiquitin chains [4] [7]. The OtUBD tool, for instance, was explicitly developed as a high-affinity UBD and has demonstrated superior performance in purifying both mono- and polyubiquitylated substrates from yeast and human cells compared to some existing methods [7].
Principle 3: Linkage and Target Bias
This principle describes the preferential enrichment of certain ubiquitin chain types or specific forms of the target protein.
- Linkage Bias in Antibodies: A significant advantage of antibody-based methods is the availability of linkage-specific reagents. Researchers can selectively enrich for proteins modified with K48-linked, K63-linked, or other specific chain types to study their unique biological functions [4]. However, general anti-ubiquitin antibodies may still exhibit inherent, and often uncharacterized, preferences for certain chain types.
- Linkage and Form Bias in UBDs: UBD-based tools like TUBEs generally exhibit a strong preference for polyubiquitinated proteins over monoubiquitinated ones, due to their reliance on multivalent interactions [7]. Their efficiency can also vary depending on the chain linkage type and length. In contrast, the OtUBD tool has been reported to efficiently detect monoubiquitylation and ubiquitin linkages to non-canonical sites, a specific advantage over some other methods [7].
Comparative Performance Analysis
The following tables summarize the key characteristics and quantitative performance data of antibody-based and UBD-based enrichment methods, drawing from direct comparisons in the literature.
Table 1: Qualitative Comparison of Enrichment Methodologies
| Feature | Antibody-Based Enrichment | UBD-Based Enrichment (e.g., TUBEs) | OtUBD-Based Enrichment |
|---|---|---|---|
| Basis of Recognition | Molecular recognition of specific epitopes | Recognition of the ubiquitin fold | High-affinity recognition of ubiquitin fold |
| Target Specificity | High for specific epitopes; can be linkage-specific | Broad for polyubiquitin chains; linkage-preference possible | Broad for both mono- and polyubiquitin |
| Typical Affinity | Nanomolar (monoclonal antibodies) | Micromolar (single UBD), enhanced by avidity | High affinity (characterized as such) |
| Advantages | Linkage-specific options available; well-established | Protects chains from DUBs; good for polyUb enrichment | Efficient for monoubiquitination; works on non-canonical sites |
| Disadvantages | Potential for steric hindrance; high cost; can have linkage bias | Low affinity for monoubiquitination; can have linkage bias | Relatively new tool; less established in diverse models |
Table 2: Experimental Performance Data from Comparative Studies
| Enrichment Method | Reported Performance Metric | Experimental Context | Source |
|---|---|---|---|
| MERT (Multiepitope) | Binding capacity 10-100x higher than mono-epitope/Fc-specific methods | Antibody/ADC enrichment from serum | [6] |
| OtUBD | Efficient detection/purification of monoubiquitylated substrates | Comparison with other methods in yeast & HeLa cells | [7] |
| OtUBD | Efficient enrichment of non-canonical ubiquitin linkages | Proof-of-principle profiling in yeast | [7] |
| TUBEs | High avidity for polyubiquitin chains; low efficiency for monoubiquitination | General methodology description | [4] [7] |
| Linkage-Specific Antibodies | Enabled identification of K48-linked tau in Alzheimer's disease | Human tissue sample analysis | [4] |
Detailed Experimental Protocols
To ensure reproducibility, this section outlines standard protocols for key enrichment methods discussed in this guide.
Protocol 1: Enrichment of Ubiquitylated Proteins Using OtUBD Affinity Purification
This protocol is adapted from the work of Zhang et al. (2022) for using the OtUBD tool to purify ubiquitylated substrates from cell lysates [7].
- Cell Lysis and Preparation: Lyse yeast or cultured mammalian cells (e.g., HeLa) in a suitable lysis buffer (e.g., 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP-40) supplemented with protease inhibitors (e.g., 1 mM PMSF), deubiquitylase (DUB) inhibitors (e.g., 10 mM N-Ethylmaleimide (NEM)), and phosphatase inhibitors. Maintain samples at 4°C throughout.
- Clarification: Clear the lysate by high-speed centrifugation (e.g., 16,000 × g for 15 minutes) to remove insoluble debris.
- Affinity Purification: Incubate the clarified lysate with OtUBD immobilized on a suitable resin (e.g., amylose resin if OtUBD is fused to MBP) for 2-4 hours at 4°C with gentle rotation.
- Washing: Wash the resin extensively with cold lysis buffer (at least 5-10 column volumes) to remove non-specifically bound proteins.
- Elution: Elute the bound ubiquitylated proteins using a competitive elution buffer. For MBP-fused OtUBD, elution can be performed with lysis buffer containing 10-20 mM maltose. Alternatively, a low-pH elution buffer (e.g., 0.1 M glycine, pH 2.5) can be used, followed by immediate neutralization.
- Analysis: The eluate can now be analyzed by immunoblotting with anti-ubiquitin antibodies or prepared for mass spectrometry-based proteomics.
Protocol 2: Enrichment Using Linkage-Specific Antibodies
This protocol describes the use of linkage-specific antibodies for immunoprecipitation, as commonly employed in the field [4].
- Lysate Preparation: Prepare cell lysates as described in Protocol 1, Step 1, ensuring the inclusion of DUB inhibitors to preserve ubiquitin chain integrity.
- Antibody Immobilization: Covalently cross-link the linkage-specific antibody (e.g., anti-K48 or anti-K63 ubiquitin chain) to Protein A/G beads using a cross-linker like dimethyl pimelimidate (DMP). This step prevents the co-elution of antibody heavy and light chains during downstream mass spectrometry, which can interfere with analysis.
- Immunoprecipitation: Incubate the clarified cell lysate with the antibody-coupled beads for 2-4 hours or overnight at 4°C with gentle rotation.
- Washing: Wash the beads stringently with cold lysis buffer to reduce background contamination.
- Elution: Elute the bound ubiquitylated proteins using a mild denaturing eluent suitable for downstream applications. A common choice is a solution of 1-2% SDS in Tris buffer, or a 0.1-0.2 M Glycine solution at pH 2.5 followed by neutralization.
- Analysis: Proceed with immunoblotting to validate the enrichment of specific chain types, or with tryptic digestion and LC-MS/MS for proteomic analysis of ubiquitination sites.
Visualization of Experimental Workflows
The following diagram illustrates the key decision-making pathways and procedural steps for selecting and implementing these enrichment strategies, based on the research objectives.
Decision Workflow for Ubiquitin Enrichment Methods
The Scientist's Toolkit: Key Research Reagents
The following table catalogues essential reagents and tools used in the featured enrichment methodologies.
Table 3: Essential Reagents for Ubiquitin Enrichment Studies
| Reagent / Tool | Function in Enrichment | Example Use Case |
|---|---|---|
| Linkage-Specific Antibodies | Immunoprecipitation of proteins modified with a specific ubiquitin chain type (K48, K63, etc.). | Studying proteasomal targeting via K48-linkages [4]. |
| TUBEs (Tandem UBDs) | High-avidity capture of polyubiquitinated proteins; protection from DUBs. | Global profiling of the polyubiquitylome; stabilizing labile ubiquitination events [4] [7]. |
| OtUBD Affinity Resin | High-affinity purification of both mono- and polyubiquitylated substrates. | Comprehensive ubiquitylome profiling, including monoubiquitylation [7]. |
| DUB Inhibitors (e.g., NEM) | Preserve the ubiquitin signal by inhibiting deubiquitylating enzymes during sample prep. | Essential additive in lysis buffer for all ubiquitin enrichment protocols [7]. |
| diGly Remnant Antibodies | Enrich for tryptic peptides containing the GlyGly-Lys remnant; identifies ubiquitination sites via MS. | Bottom-up proteomics to map ubiquitination sites at the system-wide level [7]. |
Protein ubiquitination is a pivotal post-translational modification that regulates virtually all aspects of eukaryotic cell biology, governing processes from protein degradation to DNA repair and immune signaling [4] [8]. The ubiquitin (Ub) system exhibits remarkable complexity—Ub can be attached to substrates as a single molecule (monoubiquitination) or form polyubiquitin chains through one of twelve identified linkage types (eight amide linkages and four ester linkages) [9]. These linkage types include the well-characterized K48-linked chains that target substrates for proteasomal degradation and K63-linked chains involved in non-degradative signaling, along with less understood "atypical" linkages (K6, K11, K27, K29, K33, M1) [4] [9]. This vast array of structurally and functionally distinct modifications constitutes what is known as the "Ubiquitin Code" [9].
Deciphering this code requires analytical tools capable of capturing and identifying ubiquitinated proteins from complex biological samples. However, the low stoichiometry of ubiquitination, dynamic nature of modifications, and vast structural diversity present significant technical challenges [4]. Traditional antibody-based methods have limitations including linkage bias, high cost, and variable affinity [4] [10]. In response to these challenges, Ubiquitin-Binding Domain (UBD)-based enrichment approaches have emerged as powerful alternatives that leverage nature's own ubiquitin recognition mechanisms to achieve high-affinity, broad-specificity capture of ubiquitinated proteins [4] [11]. This guide provides a comprehensive comparison of UBD-based enrichment strategies, focusing on their core principles of ubiquitin chain recognition, binding affinity, and unbiased capture relative to alternative methodologies.
Fundamental Principles of UBD-Based Enrichment
Ubiquitin-binding domains (UBDs) are modular protein domains that non-covalently interact with ubiquitin, serving as natural recognition modules in cellular ubiquitin signaling pathways [12]. Approximately 20 distinct UBD families have been identified in the human genome, including UBA (Ubiquitin-Associated), UIM (Ubiquitin-Interacting Motif), and NZF (Npl4 Zinc Finger) domains [12] [9]. These domains typically exhibit low to moderate affinity for ubiquitin when isolated (micromolar range), which allows for reversible interactions necessary for dynamic cellular signaling [12].
The core innovation in UBD-based enrichment technologies involves engineering these natural recognition domains to overcome their inherent affinity limitations while preserving their linkage recognition properties. Two principal engineering strategies have been employed:
- Tandem UBD Constructs: Multiple UBDs are connected via flexible linkers, significantly increasing avidity through multivalent interactions [4] [11].
- Hybrid UBD Designs: UBDs with different linkage preferences and binding modes are combined to create constructs with broad specificity across multiple ubiquitin chain types [11].
These engineered UBDs serve as affinity reagents that can be coupled to solid supports for enriching ubiquitinated proteins and peptides from complex mixtures, enabling subsequent identification and analysis by western blotting or mass spectrometry [4] [10] [11].
Comparative Performance Analysis: UBD-Based vs. Alternative Enrichment Methods
Multiple strategies exist for enriching ubiquitinated proteins, each with distinct advantages and limitations. The table below provides a comprehensive comparison of the three primary enrichment approaches.
Table 1: Comparison of Major Ubiquitinated Protein Enrichment Methodologies
| Method | Mechanism | Affinity/Sensitivity | Linkage Bias | Key Advantages | Major Limitations |
|---|---|---|---|---|---|
| UBD-Based | Engineered tandem UBDs (e.g., ThUBD) | 16-fold wider linear range vs. TUBE; captures 0.625μg ubiquitinated proteins [10] | Minimal bias; binds all 7 lysine-linked chains [11] | High affinity; preserves native ubiquitination; suitable for tissues [4] [10] | Requires protein engineering; optimization needed for different formats [11] |
| Antibody-Based | Immunoaffinity with anti-ubiquitin antibodies | Variable; limited by antibody affinity and epitope accessibility [4] | Significant bias with most conventional antibodies [4] [9] | Works with native ubiquitin; established protocols [4] | High cost; linkage specificity requires multiple antibodies; non-specific binding [4] |
| Tag-Based | Expression of epitope-tagged ubiquitin (e.g., His, HA, Strep) | Limited by tag accessibility; lower identification efficiency [4] | Depends on tagged ubiquitin incorporation | Easy implementation; widely accessible [4] | Genetic manipulation required; artifacts from tag; not suitable for human tissues [4] |
Quantitative Performance Comparison of UBD Technologies
Recent technological advances have produced increasingly sophisticated UBD constructs with remarkable performance characteristics. The following table summarizes the experimental performance data for leading UBD-based technologies.
Table 2: Performance Characteristics of Engineered UBD Constructs
| UBD Construct | Composition | Affinity Improvement | Linkage Coverage | Documented Applications |
|---|---|---|---|---|
| ThUBD (Tandem Hybrid UBD) | DSK2p-UBA + Ubiquilin2-UBA or DSK2p-UBA + A20-ZnF [11] | Markedly higher than natural UBDs [11] | All 7 lysine-linked chains with almost unbiased high affinity [11] | Identified 1,125 ubiquitinated proteins with modification sites from mammalian cells [11] |
| ThUBD-coated plates | Engineered ThUBD on high-density 96-well plates [10] | 16-fold wider linear range vs. TUBE technology; binds ~5pmol polyUb chains [10] | Unbiased enrichment across all ubiquitin chain types [10] | High-throughput detection; PROTAC drug development; dynamic ubiquitination monitoring [10] |
| TUBE (Tandem Ubiquitin Binding Entity) | Multiple UBA domains in tandem [10] | Lower affinity compared to ThUBD [10] | Noticeable bias toward specific linkage types [10] | Historical use in ubiquitin proteomics; being superseded by higher-affinity alternatives [10] |
Experimental Validation and Protocol Details
The superior performance of engineered ThUBDs has been demonstrated through multiple rigorous experimental approaches. In one foundational study, researchers systematically evaluated the affinity of various UBDs to different ubiquitin chain types, selecting UBDs with high affinity and evaluating various combinations to construct two artificial tandem hybrid UBDs (ThUDQ2 and ThUDA20) [11]. The key experimental steps included:
- UBD Screening: Multiple natural UBDs were screened for affinity to different ubiquitin linkage types using surface plasmon resonance (SPR) and binding assays [11].
- Construct Design: Selected UBDs were combined in tandem arrangements with optimized linkers to create hybrid constructs [11].
- Affinity Measurement: Binding affinity was quantified using SPR, demonstrating markedly higher affinity compared to naturally occurring UBDs [11].
- Linkage Specificity Profiling: Specificity was assessed against all seven lysine-linked ubiquitin chains, showing nearly unbiased high affinity [11].
- Proteomic Application: ThUBD-based profiling with mass spectrometry identified 1,092 and 7,487 putative ubiquitinated proteins from yeast and mammalian cells, respectively, with 362 and 1,125 proteins having identified ubiquitination sites [11].
For the ThUBD-coated plate technology, the experimental protocol involved:
- Coating 1.03 μg ± 0.002 of ThUBD on Corning 3603-type 96-well plates [10]
- Systematic optimization of coating conditions, washing buffers, and detection conditions [10]
- Validation through comparison with TUBE-coated plates, demonstrating 16-fold wider linear range for capturing polyubiquitinated proteins [10]
- Application to global ubiquitination profiles and target-specific ubiquitination status [10]
Technical Implementation and Research Reagent Solutions
Essential Research Reagents for UBD-Based Enrichment
Successful implementation of UBD-based enrichment requires specific reagents and materials. The following table details key components of the UBD research toolkit.
Table 3: Essential Research Reagent Solutions for UBD-Based Enrichment
| Reagent/Resource | Function/Purpose | Specific Examples |
|---|---|---|
| Engineered UBD Constructs | High-affinity capture of ubiquitinated proteins | ThUBD (ThUDQ2, ThUDA20) [11] |
| Immobilization Supports | Solid-phase presentation of UBDs | Ni-NTA agarose (His-tagged UBDs); Strep-Tactin (Strep-tagged UBDs); high-density 96-well plates [10] [11] |
| Binding and Wash Buffers | Optimize specific binding and reduce non-specific interactions | Systematically optimized buffers for ThUBD-coated plates [10] |
| Detection Reagents | Identify and quantify captured ubiquitinated proteins | ThUBD-HRP conjugates; linkage-specific antibodies for validation [10] |
| Ubiquitin Chain Standards | Method validation and quantification | Recombinant Ub-GFP, Ub2-GFP, Ub4-GFP [10] |
| Mass Spectrometry Compatibility | Identification of ubiquitination sites | Compatibility with tryptic digestion and LC-MS/MS analysis [11] |
Practical Implementation Workflow
The typical workflow for UBD-based enrichment involves several key stages:
- Sample Preparation: Cell lysis under non-denaturing conditions to preserve ubiquitin modifications
- UBD Immobilization: Coupling of engineered UBD constructs to appropriate solid supports
- Affinity Enrichment: Incubation of cell lysates with UBD-conjugated supports
- Stringent Washing: Removal of non-specifically bound proteins
- Elution and Analysis: Recovery of ubiquitinated proteins for downstream applications
This workflow has been successfully applied to both global ubiquitin proteomics and targeted studies of specific proteins of interest [10] [11].
Applications in Drug Discovery and Development
UBD-based technologies have found particularly valuable applications in pharmaceutical development, especially in the rapidly advancing field of targeted protein degradation. The high-throughput capabilities of ThUBD-coated plates enable rapid screening and optimization of Proteolysis-Targeting Chimeras (PROTACs) and other molecular degraders [10]. These compounds redirect E3 ubiquitin ligases to target specific proteins of interest for ubiquitination and degradation, making precise monitoring of ubiquitination status essential for their development [10].
The unbiased capture capability of engineered ThUBDs is particularly valuable for understanding the mechanism of action of these novel therapeutic modalities, as different ubiquitin chain types can influence the efficiency and outcomes of targeted protein degradation [10]. Furthermore, the ability to monitor dynamic changes in ubiquitination in response to treatment provides crucial insights for lead optimization and candidate selection in drug discovery pipelines [10].
UBD-based enrichment strategies represent a significant advancement in our ability to decipher the complex ubiquitin code. Through strategic protein engineering, these approaches overcome the inherent limitations of natural UBDs while preserving their biological relevance. The development of tandem hybrid UBDs with enhanced affinity and minimal linkage bias provides researchers with powerful tools for comprehensive ubiquitin proteomics, drug discovery, and fundamental mechanistic studies of ubiquitin signaling.
As the ubiquitin field continues to evolve, further refinements in UBD design and implementation will undoubtedly enhance our understanding of this essential regulatory system and facilitate the development of novel therapeutics targeting the ubiquitin-proteasome system.
Visual Appendix
Ubiquitin Code Complexity and Detection Challenge
UBD Engineering Strategy for Enhanced Enrichment
The Ubiquitin-Proteasome System (UPS) and Why Efficient Enrichment is Crucial for Its Study
The Ubiquitin-Proteasome System (UPS) is a critical regulatory mechanism in eukaryotic cells, responsible for the controlled degradation of intracellular proteins. This intricate system governs nearly every cellular process, from cell cycle progression and DNA repair to immune responses and signal transduction. The study of ubiquitination—the process by which ubiquitin is attached to target proteins—is fraught with technical challenges, primarily due to the low stoichiometry of modified proteins and the complexity of ubiquitin chain architectures. Efficient enrichment of ubiquitinated proteins is therefore not merely a preliminary step but a fundamental prerequisite for generating meaningful biological insights. This guide objectively compares the two dominant enrichment methodologies—antibody-based and ubiquitin-binding domain (UBD)-based approaches—evaluating their performance in the context of modern proteomic research.
The Central Role of the UPS and the Imperative for Efficient Enrichment
The UPS operates through a sequential enzymatic cascade involving E1 (activating), E2 (conjugating), and E3 (ligating) enzymes, which culminate in the covalent attachment of ubiquitin to lysine residues on substrate proteins. The 26S proteasome then recognizes and degrades these tagged proteins, but ubiquitination also serves non-proteolytic functions, influencing protein activity, localization, and interactions. The system's complexity is magnified by the ability of ubiquitin itself to form polymer chains through its own lysine residues, with different chain linkages (e.g., K48, K63, M1) dictating distinct cellular fates.
Efficient enrichment is crucial because:
- Low Abundance: The stoichiometry of ubiquitination for any given protein is typically very low under physiological conditions.
- Signal Dilution: Ubiquitinated peptides are dwarfed by their non-modified counterparts in a total proteome digest, making direct detection impossible without enrichment.
- Functional Specificity: To understand the biological consequences of ubiquitination, researchers must accurately identify not only the modified protein but also the specific site of modification and the topology of the ubiquitin chain.
Comparative Analysis of Enrichment Methodologies
The two primary strategies for isolating ubiquitinated proteins are antibody-based and UBD-based enrichment. The table below summarizes their core characteristics and performance metrics.
Table 1: Core Characteristics of Ubiquitin Enrichment Methodologies
| Feature | Antibody-Based Enrichment | UBD-Based Enrichment |
|---|---|---|
| Basis of Recognition | Immunoaffinity for the di-glycine (K-ε-GG) remnant left on trypsinized peptides [13] [14] | High-affinity protein-protein interaction with ubiquitin chains on intact proteins [15] |
| Key Technology | Anti-K-ε-GG monoclonal antibodies [15] | Tandem Hybrid Ubiquitin Binding Domains (ThUBDs) [10] |
| Typical Application | Mass spectrometry-based ubiquitin proteomics [13] [14] | Detection of endogenous ubiquitin signals in cells and tissues; western blotting [15] [10] |
| Linkage Bias | Linkage-specific antibodies exist, but general antibodies may have inherent preferences [15] | Engineered ThUBDs demonstrate unbiased recognition across all ubiquitin chain types [10] |
| Compatibility | Requires tryptic digestion; incompatible with intact protein analysis | Compatible with intact proteins and complex proteomes; suitable for functional studies |
Recent technological advancements have yielded significant performance improvements, particularly in UBD-based platforms. The following table compares quantitative data from recent studies highlighting these gains.
Table 2: Quantitative Performance Comparison of Modern Enrichment Tools
| Method / Tool | Reported Performance Metric | Comparative Outcome | Source / Context |
|---|---|---|---|
| ThUBD-coated plates | Detection sensitivity as low as 0.625 μg of proteome sample [10] | 16-fold wider linear range and higher sensitivity than TUBE-coated plates [10] | High-throughput plate-based assay |
| UbiFast (On-antibody TMT) | Quantification of >10,000 ubiquitylation sites from 500 μg of peptide per sample [14] | Enabled highly multiplexed, deep-scale profiling from limited tissue samples [14] | Mass spectrometry proteomics |
| On-antibody TMT labeling | Relative yield of 85.7% K-ε-GG peptides [14] | Nearly double the relative yield (44.2%) of in-solution labeling methods [14] | Sample preparation for mass spectrometry |
Detailed Experimental Protocols
Protocol 1: UbiFast - On-Antibody TMT Labeling for Mass Spectrometry
This protocol allows for multiplexed, quantitative analysis of thousands of ubiquitination sites from small amounts of sample, ideal for clinical or primary tissue research [14].
- Sample Preparation: Lyse cells or tissue in a denaturing buffer (e.g., 8M Urea). Reduce, alkylate, and digest the proteins into peptides using trypsin. This digestion generates the characteristic K-ε-GG remnant on modified lysines.
- Peptide Enrichment: Incubate the peptide mixture (e.g., 0.5-1 mg) with anti-K-ε-GG antibody conjugated to beads. Wash thoroughly to remove non-specifically bound peptides.
- On-Bead TMT Labeling: While the K-ε-GG peptides are bound to the antibody, resuspend the beads in a solution containing Tandem Mass Tag (TMT) reagents. The TMT labels the N-termini and lysine side chains of the enriched peptides, but the K-ε-GG remnant itself is protected from labeling. Quench the reaction with hydroxylamine [14].
- Peptide Elution and Pooling: Elute the TMT-labeled peptides from the antibody. Combine (multiplex) the samples from different experimental conditions.
- LC-MS/MS Analysis: Desalt the pooled sample and analyze by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The TMT tags enable simultaneous quantification across all conditions.
The workflow is illustrated below.
Protocol 2: ThUBD-Coated Plate Assay for High-Throughput Analysis
This protocol is designed for rapidly quantifying global or target-specific ubiquitination levels, with applications in drug discovery like PROTAC development [10].
- Plate Coating: Coat high-binding 96-well plates with the recombinant ThUBD protein (approximately 1 μg per well) to create a capture surface.
- Sample Application: Add the protein sample of interest (e.g., cell lysate) to the ThUBD-coated wells. Incubate to allow ubiquitinated proteins to bind.
- Stringent Washing: Wash the wells with a specialized buffer to remove all non-specifically bound material.
- Detection: Detect the captured ubiquitinated proteins using a primary antibody against your target protein (for target-specific analysis) or an anti-ubiquitin antibody (for global ubiquitination profiling), followed by an HRP-conjugated secondary antibody. Quantify the signal via chemiluminescence.
The high-throughput process is summarized in the following diagram.
The Scientist's Toolkit: Essential Research Reagents
Successful ubiquitination studies depend on specific, high-quality reagents. The table below details key solutions for the experiments described.
Table 3: Key Research Reagent Solutions for UPS Enrichment Studies
| Reagent / Tool | Primary Function | Application Context |
|---|---|---|
| Anti-K-ε-GG Antibody | Immunoaffinity enrichment of tryptic peptides containing the di-glycine ubiquitin remnant [13] [15]. | Mass spectrometry-based ubiquitin proteomics (e.g., UbiFast protocol). |
| Tandem Mass Tag (TMT) | Isobaric chemical labels for multiplexed, relative quantification of peptides across multiple samples in a single MS run [14]. | Quantitative proteomics to compare ubiquitination sites under different conditions. |
| Tandem Hybrid UBD (ThUBD) | Engineered high-affinity binding domain for unbiased capture of polyubiquitinated proteins, regardless of chain linkage type [10]. | Plate-based assays and western blotting to assess global ubiquitination or specific protein ubiquitination. |
| Linkage-Specific Ub Antibodies | Antibodies that recognize a particular ubiquitin chain topology (e.g., K48-only, K63-only) [15]. | Immunoblotting to determine the functional fate (e.g., degradation signal) of a ubiquitinated protein. |
| N-Ethylmaleimide (NEM) | Cysteine protease inhibitor that is added to lysis buffers to irreversibly inhibit deubiquitinating enzymes (DUBs) [13]. | Essential for all protocols to preserve the native ubiquitination state of proteins during sample preparation. |
The choice between antibody-based and UBD-based enrichment methods is not a matter of declaring one universally superior, but of aligning tool capability with experimental goal. Antibody-based methods, particularly when coupled with advanced workflows like on-bead TMT labeling, provide unparalleled depth and precision for site-specific ubiquitin proteomics. In contrast, the latest UBD-based technologies, exemplified by ThUBD-coated plates, offer superior performance in affinity, linkage neutrality, and throughput for functional, intact-protein analyses. As the UPS continues to emerge as a therapeutic target in cancer and neurodegenerative diseases, the strategic selection and continued refinement of these enrichment tools will be fundamental to cracking the molecular mechanisms of disease and advancing novel therapeutics like PROTACs.
The ubiquitin-proteasome system (UPS) serves as the primary proteolytic pathway for regulated protein degradation in eukaryotic cells, tightly governing critical cellular processes including DNA repair, stress response, and cell proliferation [16]. This system functions through a coordinated enzymatic cascade: ubiquitin-activating enzymes (E1) activate ubiquitin, ubiquitin-conjugating enzymes (E2) transfer the activated ubiquitin, and ubiquitin ligases (E3) facilitate the final attachment of ubiquitin to specific substrate proteins [16] [4]. Once a substrate is tagged with a polyubiquitin chain (typically at least four ubiquitin molecules), it is recognized and hydrolyzed by the 26S proteasome [16]. The versatility of ubiquitination extends beyond mere degradation; it regulates diverse fundamental features of protein substrates, including stability, activity, and localization, with dysregulation of this process leading to many pathologies, including cancer and neurodegenerative diseases [4].
The study of protein ubiquitination presents significant technical challenges due to the low stoichiometry of modified proteins, the complexity of ubiquitin chain architectures (homotypic, heterotypic, and branched chains), and the transient nature of the modifications [4]. To overcome these challenges, researchers have developed various enrichment strategies, primarily falling into two categories: antibody-based methods and ubiquitin-binding domain (UBD)-based methods. This guide provides an objective comparison of these approaches, focusing on their application in basic research and the development of revolutionary technologies like Proteolysis Targeting Chimeras (PROTACs).
Methodological Comparison: UBD-Based vs. Antibody-Based Enrichment
The accurate profiling of ubiquitinated proteins requires efficient enrichment from complex biological samples to avoid interference from non-ubiquitinated proteins. The following sections detail the core methodologies, and Table 1 provides a structured comparison of their performance characteristics.
Ubiquitin-Binding Domain (UBD)-Based Approaches
UBD-based approaches utilize proteins or protein domains that naturally recognize and bind to ubiquitin. These domains are found in various proteins, including some E3 ubiquitin ligases, deubiquitinating enzymes (DUBs), and ubiquitin receptors [4]. A significant advancement in this area is the development of engineered tandem hybrid UBDs (ThUBDs). For instance, researchers have systematically evaluated UBD affinity and constructed artificial tandems like ThUDQ2 (comprising DSK2p-derived UBA and ubiquilin 2-derived UBA) and ThUDA20 (made from DSK2p-derived UBA and RABGEF1-derived A20-ZnF) [11]. These ThUBDs exhibit markedly higher affinity to ubiquitinated proteins compared to naturally occurring single UBDs and display almost unbiased high affinity to all seven lysine-linked ubiquitin chains [11].
Protocol for ThUBD-Based Enrichment: [11]
- Construct Design: Engineer the DNA sequence for the chosen ThUBD (e.g., ThUDQ2 or ThUDA20) into an appropriate expression vector.
- Protein Expression: Express the recombinant ThUBD protein in a suitable host system, such as E. coli.
- Affinity Resin Preparation: Immobilize the purified ThUBD protein onto a solid-phase resin (e.g., glutathione-sepharose for GST-tagged ThUBDs).
- Cell Lysis: Lyse cells or tissues using a non-denaturing lysis buffer to preserve protein-protein interactions.
- Enrichment: Incubate the cell lysate with the ThUBD-bound resin. The ThUBD will bind ubiquitinated proteins with high affinity.
- Washing: Wash the resin thoroughly with lysis buffer to remove non-specifically bound proteins.
- Elution: Elute the enriched ubiquitinated proteins using a denaturing buffer (e.g., containing SDS) or a competitive elution with free ubiquitin.
- Downstream Analysis: The eluate can then be analyzed by immunoblotting or prepared for mass spectrometry-based proteomics.
Antibody-Based Approaches
Antibody-based methods rely on immunoenrichment using antibodies that recognize ubiquitin. These can be pan-specific anti-ubiquitin antibodies (e.g., P4D1, FK1, FK2) that recognize all ubiquitin linkages or linkage-specific antibodies (e.g., K48-, K63-specific) that target particular polyubiquitin chain architectures [4]. This approach allows for the profiling of endogenously ubiquitinated substrates without the need for genetic manipulation, making it feasible for use with animal tissues or clinical samples [4].
Protocol for Antibody-Based Enrichment: [4]
- Cell Lysis: Lyse cells or homogenize tissues in a denaturing lysis buffer (e.g., containing SDS) to inactivate deubiquitinases and preserve the ubiquitination state.
- Dilution: Dilute the lysate significantly with a non-denaturing buffer to reduce the concentration of SDS below a level that would interfere with antibody binding.
- Pre-clearing: Incubate the diluted lysate with control IgG and protein A/G beads to reduce non-specific binding.
- Immunoprecipitation: Incubate the pre-cleared lysate with the specific anti-ubiquitin antibody (pan-specific or linkage-specific) conjugated to protein A/G beads.
- Washing: Wash the beads extensively with a suitable wash buffer to remove unbound proteins.
- Elution: Elute the ubiquitinated proteins using a low-pH buffer or Laemmli sample buffer for direct downstream analysis by immunoblotting or mass spectrometry.
Table 1: Performance Comparison of Ubiquitinated Protein Enrichment Methods
| Feature | UBD-Based Methods (incl. ThUBD) | Antibody-Based Methods |
|---|---|---|
| Basis of Enrichment | Affinity of engineered/ natural ubiquitin-binding domains [11] | Immunorecognition by anti-ubiquitin antibodies [4] |
| Key Tools/Reagents | ThUBD proteins (ThUDQ2, ThUDA20), affinity resins (e.g., glutathione-sepharose) [11] | Pan-specific (P4D1, FK2) or linkage-specific anti-ubiquitin antibodies [4] |
| Affinity & Efficiency | Very high affinity with engineered ThUBDs; high efficiency in unbiased binding to various linkages [11] | High affinity, but dependent on antibody quality and epitope accessibility [4] |
| Linkage Specificity | Broad specificity (ThUBDs show high affinity for all 7 Lys-linked chains); can be tailored [11] | Can be broad (pan-specific) or highly selective (linkage-specific) [4] |
| Applicability to Endogenous Proteins | Yes, effective for endogenous proteins [11] | Yes, the primary strength; no genetic tags needed [4] |
| Throughput & Scalability | High, suitable for proteomic-scale studies [11] | Moderate, can be limited by antibody cost and availability [4] |
| Major Advantages | High, largely unbiased affinity; cost-effective for large-scale studies; tunable [11] | Direct application to any sample, including clinical tissues; linkage-specific analysis [4] |
| Major Limitations | Requires recombinant protein production; potential for non-specific binding if not optimized | High cost of quality antibodies; potential for non-specific binding (e.g., to antibody beads) [4] |
| Typical Identifications (Proteomics) | ~7,500 putative ubiquitinated proteins from mammalian cells [11] | Lower identification numbers in published studies (e.g., 96 ubiquitination sites) [4] |
Application in Targeted Protein Degradation: The Rise of PROTACs
The understanding of the UPS has directly enabled a paradigm shift in drug discovery: Targeted Protein Degradation (TPD). Proteolysis Targeting Chimeras (PROTACs) are the most advanced modality in this field [17]. These are heterobifunctional molecules that consist of three parts: a ligand that binds a target protein of interest (POI), a ligand that recruits an E3 ubiquitin ligase, and a linker connecting the two [16] [17]. The mechanism of action is illustrated in the diagram below.
Figure 1: Mechanism of PROTAC-Induced Protein Degradation.
PROTACs offer several advantages over traditional small-molecule inhibitors [16] [17]:
- Event-Driven Catalysis: They operate sub-stoichiometrically, as a single PROTAC molecule can be recycled to degrade multiple copies of the target protein.
- Targeting "Undruggable" Proteins: They can degrade proteins that lack a defined active site (e.g., transcription factors, scaffolding proteins) by relying on binding, not inhibition.
- Overcoming Resistance: They can effectively degrade proteins that have developed resistance to inhibitors through mutations or overexpression.
The clinical pipeline for PROTACs has expanded rapidly. As of 2025, over 40 PROTAC drug candidates are in clinical trials, with three having advanced to Phase III studies, as detailed in Table 2 [18] [17].
Table 2: Selected PROTAC Degraders in Clinical Trials (2025 Update)
| Drug Candidate | Company(s) | Target | Indication(s) | Development Status (as of 2025) |
|---|---|---|---|---|
| Vepdegestrant (ARV-471) | Arvinas / Pfizer | Estrogen Receptor (ER) | ER+/HER2- Breast Cancer | Phase III (Fast Track designation) [18] |
| BMS-986365 (CC-94676) | Bristol Myers Squibb (BMS) | Androgen Receptor (AR) | Metastatic Castration-Resistant Prostate Cancer (mCRPC) | Phase III [18] |
| BGB-16673 | BeiGene | Bruton's Tyrosine Kinase (BTK) | Relapsed/Refractory B-cell Malignancies | Phase III [18] |
| ARV-110 | Arvinas | Androgen Receptor (AR) | mCRPC | Phase II [18] |
| KT-474 (SAR444656) | Kymera | IRAK4 | Hidradenitis Suppurativa & Atopic Dermatitis | Phase II [18] |
A key challenge in PROTAC development is the limited repertoire of E3 ligases used in current clinical candidates, with most relying on ligands for CRBN or VHL [19]. Expanding the E3 ligase toolbox is an active area of research, with efforts focused on recruiting other ligases like MDM2, IAP, DCAF16, and KEAP1 to enable tissue-specific targeting and degrade a wider array of proteins [17] [19].
The Scientist's Toolkit: Essential Research Reagents
The following table lists key reagents and materials essential for conducting research in protein ubiquitination and PROTAC development.
Table 3: Essential Research Reagents and Solutions
| Reagent / Solution | Function / Application | Key Considerations |
|---|---|---|
| Tagged Ubiquitin (His-, Strep-) | Affinity purification of ubiquitinated proteins from cell lysates for proteomic analysis [4]. | May alter Ub structure; can co-purify endogenous biotinylated/His-rich proteins [4]. |
| Pan-Ubiquitin Antibodies (P4D1, FK1/FK2) | Immunoprecipitation and immunoblotting for total ubiquitinated proteins [4]. | Quality and specificity vary between lots; high cost for large-scale use [4]. |
| Linkage-Specific Ub Antibodies | Enrichment and detection of specific polyubiquitin chain linkages (e.g., K48, K63) [4]. | Essential for determining the functional consequence of ubiquitination. |
| Engineered Tandem UBDs (ThUBDs) | High-efficiency, broad-spectrum enrichment of ubiquitinated proteins for proteomics [11]. | Requires production of recombinant protein; optimization of binding/wash conditions. |
| E3 Ligase Ligands (e.g., for CRBN, VHL) | Key components for designing and synthesizing PROTAC molecules [16] [17]. | Determines the E3 ligase recruited and influences PROTAC efficiency and selectivity. |
| PROTAC Target Ligands | Binds the protein of interest (POI) in a PROTAC molecule (e.g., AR/ER ligands, kinase inhibitors) [16]. | The starting point for PROTAC design; affinity influences ternary complex formation. |
| Linker Chemistry | Connects the E3 ligand and POI ligand in a PROTAC; influences physicochemical properties and ternary complex geometry [16] [17]. | Length, composition, and rigidity are critical optimization parameters. |
The methods for enriching and studying ubiquitinated proteins, particularly the advanced UBD-based and antibody-based techniques, are foundational to modern biomedicine. The quantitative comparison provided in this guide highlights that while UBD-based methods like engineered ThUBDs offer superior affinity and are highly suited for discovery-stage proteomics, antibody-based methods remain indispensable for studying endogenous ubiquitination in pathophysiological contexts. The deep understanding of the UPS provided by these tools has directly catalyzed the development of transformative therapeutic modalities like PROTACs, which are now demonstrating significant clinical promise. The continued refinement of enrichment methodologies and the expansion of the E3 ligase toolbox will undoubtedly accelerate the development of next-generation degraders, further bridging the gap from basic research to innovative drug development.
From Theory to Bench: Standard Protocols for UBD and Antibody Enrichment
Immunoprecipitation (IP) serves as a cornerstone technique for isolating ubiquitinated proteins from complex biological samples, enabling researchers to study this crucial post-translational modification. Ubiquitination is a versatile modification that regulates fundamental features of protein substrates, including stability, activity, and localization [4]. The technique leverages the highly specific interaction between anti-ubiquitin antibodies and their target proteins to selectively capture ubiquitin-protein conjugates from cell or tissue lysates [20]. Within the context of evaluating enrichment efficiency, two primary methodological approaches have emerged: antibody-based enrichment using anti-ubiquitin antibodies and Ubiquitin Binding Domain (UBD)-based approaches that utilize proteins containing ubiquitin-binding domains to capture ubiquitinated proteins [4]. Each method presents distinct advantages and limitations in specificity, yield, and applicability to different research scenarios. This guide provides a detailed, step-by-step protocol for performing immunoprecipitation with anti-ubiquitin antibodies, while objectively comparing its performance against alternative UBD-based methods to help researchers select the most appropriate approach for their experimental needs.
Key Reagents and Materials
The Scientist's Toolkit: Essential Research Reagents
Table 1: Essential Reagents for Ubiquitin Immunoprecipitation
| Reagent/Material | Function/Application | Examples/Specifications |
|---|---|---|
| Anti-Ubiquitin Antibodies | Detection and enrichment of ubiquitinated proteins | Monoclonal (e.g., 14-6078-82) [21]; Polyclonal (e.g., PA1-10023, PA3-16717) [21]; Linkage-specific antibodies (K48, K63, etc.) [4] |
| Cell Lysis Buffers | Extraction of proteins while preserving ubiquitination | NP-40 Buffer (mild): 150 mM NaCl, 1% NP-40, 50 mM Tris-HCl pH 8.0 [20]; RIPA Buffer (harsh): 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS [20] |
| Protease Inhibitors | Prevent degradation of ubiquitin conjugates | Commercial cocktails (e.g., ab65621) [20] |
| Phosphatase Inhibitors | Preserve phosphorylation states | Optional additions (e.g., ab201112) [20] |
| Protein A/G Beads | Immobilization of antibody-antigen complexes | Agarose or magnetic beads coupled to Protein A or G [20] |
| Wash Buffers | Remove non-specifically bound proteins | Variations of lysis buffer with adjusted salt/detergent concentrations [20] |
| Elution Buffers | Release bound ubiquitinated proteins | Low pH buffer, SDS sample buffer, or competitive elution [20] |
Experimental Protocol: Step-by-Step Workflow
Stage 1: Lysate Preparation
Proper lysate preparation is critical for successful immunoprecipitation of ubiquitinated proteins, as it directly impacts the ability to isolate intact protein complexes [20].
Cell Harvesting and Lysis:
- Isolate cells and suspend in appropriate lysis buffer (300 µL for 1-3×10⁷ cells) [20]
- For adherent cells, detach with trypsin-based solutions, wash with PBS, and pellet cells before adding lysis buffer [20]
- For tissues, snap-freeze and homogenize using a bead beater homogenizer (200 mg tissue in 1200 µL lysis buffer) [20]
Lysis Conditions:
Protein Quantification:
Stage 2: Pre-clearing (Optional)
Pre-clearing lysates is an optional step that can help increase the purity of ubiquitinated proteins obtained by IP by reducing non-specific binding [20].
Procedure:
Considerations:
Stage 3: Immunoprecipitation
Antibody Binding:
- Add optimized concentration of anti-ubiquitin antibody to lysate
- Incubate 2-4 hours at 4°C with gentle rotation to form antibody-ubiquitin complexes
Beads Capture:
Washing:
- Pellet beads by brief centrifugation (500-1000 × g for 1-2 minutes)
- Wash 3-4 times with appropriate wash buffer (typically lysis buffer variants)
- Remove supernatant completely after each wash while avoiding bead disruption
Elution:
- Elute bound ubiquitinated proteins using low pH buffer (0.1-0.2 M glycine, pH 2.5-3.0) or SDS sample buffer for direct western blot analysis [20]
- Neutralize low pH eluates immediately with Tris buffer, pH 8.0-9.0
Method Comparison: Antibody vs. UBD-Based Enrichment
Performance Metrics and Applications
Table 2: Comparative Analysis of Ubiquitin Enrichment Methods
| Parameter | Antibody-Based Methods | UBD-Based Methods |
|---|---|---|
| Mechanism | Antigen-antibody interaction with anti-ubiquitin antibodies [21] | Ubiquitin-binding domains (UBDs) recognizing ubiquitin linkages [4] |
| Specificity | High specificity with linkage-specific antibodies (e.g., K48, K63) [4] | Variable specificity; some UBDs recognize specific linkages, others are general [4] |
| Applications | Western Blot, IHC, ICC, ELISA, Flow Cytometry [21]; enrichment for MS analysis [4] | Primarily enrichment for mass spectrometry analysis [4] |
| Throughput | Medium to high throughput with commercial antibody availability [21] | Lower throughput due to protein expression requirements [4] |
| Sample Compatibility | Compatible with cell lines, animal tissues, and clinical samples [21] [4] | Primarily cell-based systems; limited application to tissues [4] |
| Key Advantages | - Wide range of validated antibodies [21]- Linkage-specific options available [4]- Suitable for various detection methods [21] | - Captures endogenous ubiquitination without tags [4]- Can preserve native ubiquitin configurations [4] |
| Limitations | - Potential epitope masking [4]- Antibody non-specificity concerns [4]- Higher cost for quality antibodies [4] | - Lower affinity of single UBDs [4]- Requires tandem UBDs for efficient capture [4]- Limited commercial availability |
Quantitative Performance Data
Table 3: Experimental Performance Metrics from Literature
| Enrichment Method | Identified Ubiquitination Sites | Sample Type | Reference |
|---|---|---|---|
| His-Tagged Ub (Peng et al.) | 110 sites on 72 proteins | S. cerevisiae | [4] |
| StUbEx System (Akimov et al.) | 277 unique sites on 189 proteins | HeLa cells | [4] |
| Strep-Tagged Ub (Danielsen et al.) | 753 lysine ubiquitylation sites on 471 proteins | U2OS/HEK293T | [4] |
| FK2 Antibody (Denis et al.) | 96 ubiquitination sites | MCF-7 breast cancer cells | [4] |
Technical Considerations and Optimization
Ubiquitination Complexity and Detection Challenges
Ubiquitination presents unique challenges for detection and enrichment due to its remarkable complexity. Unlike simpler post-translational modifications, ubiquitin can form diverse conjugates ranging from single ubiquitin monomers to polymers with different lengths and linkage types [4]. The ubiquitin molecule itself contains one N-terminal methionine residue (M1) and seven lysine residues (K6, K11, K27, K29, K33, K48, K63) that provide eight free -NH₂ groups as linkage sites for conjugating with the C-terminus of distal ubiquitin molecules [4]. This results in homotypic chains (same linkage type), heterotypic chains (mixed linkages), and branched chains that further complicate analysis [4]. Additionally, the stoichiometry of protein ubiquitination is typically very low under normal physiological conditions, increasing the difficulty of identifying ubiquitinated substrates [4]. These factors necessitate careful method selection and rigorous optimization for successful detection and characterization of ubiquitination events.
Method Selection Guidelines
Choosing between antibody-based and UBD-based enrichment methods requires careful consideration of research goals and experimental constraints. Antibody-based approaches are particularly advantageous when working with clinical samples or animal tissues where genetic manipulation is infeasible [4]. The availability of linkage-specific antibodies (M1-, K11-, K27-, K48-, K63-linkage specific) enables researchers to study the functional consequences of specific ubiquitin chain types without requiring specialized cell lines or expression systems [4]. For example, K48-linked ubiquitin chains target substrate proteins to the 26S proteasome for degradation, while K63-linked chains regulate protein-protein interactions in pathways such as NF-κB activation and autophagy [4]. In contrast, UBD-based approaches may be preferable when studying endogenous ubiquitination without introducing tags that might alter ubiquitin structure or function [4]. However, the lower affinity of single UBDs often necessitates using tandem-repeated ubiquitin-binding domains to achieve efficient capture, which can complicate implementation [4]. For discovery-phase research aiming to identify novel ubiquitination sites across the proteome, tagged ubiquitin systems (His-tag or Strep-tag) combined with mass spectrometry analysis offer the highest sensitivity and coverage, though they may introduce artifacts by overexpressing modified ubiquitin [4].
Downstream Applications and Analysis
Following immunoprecipitation with anti-ubiquitin antibodies, researchers can pursue multiple analytical paths to extract biological insights. Western blotting remains the most common validation method, allowing confirmation of ubiquitination status for specific proteins of interest [21]. For comprehensive ubiquitome mapping, mass spectrometry-based proteomics enables high-throughput identification of ubiquitination sites and quantification of changes under different experimental conditions [4]. The diGly remnant (114.04 Da mass shift on modified lysine residues) serves as a signature for ubiquitination site identification by mass spectrometry [4]. Functional studies often combine immunoprecipitation with mechanistic experiments to elucidate the biological consequences of ubiquitination, such as protein degradation kinetics, changes in subcellular localization, or alterations in protein-protein interactions. In diagnostic and clinical applications, anti-ubiquitin antibodies have been used to detect pathological protein aggregates in neurodegenerative diseases, with studies demonstrating correlations between ubiquitin-positive lesion densities and cognitive status in Alzheimer's disease [22]. The selection of appropriate downstream applications should align with the research objectives, whether focused on mechanistic understanding of specific pathways or comprehensive profiling of ubiquitination changes in physiological or disease contexts.
Protein ubiquitination is a fundamental post-translational modification that regulates nearly all cellular processes in eukaryotes, including protein degradation, DNA repair, cell cycle progression, and immune responses [10] [23]. The dysregulation of ubiquitination pathways is intimately linked to the pathogenesis of numerous prevalent diseases, particularly cancers and neurodegenerative disorders [10]. As drug development increasingly focuses on targeting the ubiquitin-proteasome system, exemplified by the growth of Proteolysis-Targeting Chimeras (PROTACs), the demand for robust, high-throughput methods to study protein ubiquitination has never been greater [10].
Traditional methods for detecting ubiquitination signals face significant limitations. Mass spectrometry-based approaches require expensive instrumentation and large sample amounts, while antibody-based methods suffer from limited affinity and inherent bias toward specific ubiquitin chain types [10]. The Tandem Ubiquitin Binding Entity (TUBE) technology represented an advancement by enabling higher-throughput analysis, but its constrained affinity for ubiquitin chains and linkage bias continued to restrict detection sensitivity and accuracy [10].
This guide provides a comprehensive comparative analysis of two pivotal technologies in ubiquitin research: the novel Tandem Hybrid Ubiquitin Binding Domain (ThUBD)-coated plates and established TUBE-based methods. We objectively evaluate their performance through experimental data, detail optimized protocols, and contextualize their applications within the broader landscape of enrichment methodologies for ubiquitination research.
Fundamental Technology and Design Principles
The core distinction between these platforms lies in their molecular design and resulting ubiquitin-binding properties. TUBE (Tandem Ubiquitin Binding Entity) technology has served as a valuable tool for ubiquitination studies but exhibits inherent limitations. Its constrained affinity for ubiquitin chains and bias toward specific ubiquitin linkage types can lead to incomplete profiling of the ubiquitinome [10].
The ThUBD (Tandem Hybrid Ubiquitin Binding Domain) platform represents a significant engineering advancement. This technology employs a fusion protein that combines the advantages of different ubiquitin-binding domains, creating a reagent with superior affinity for polyubiquitinated proteins and, crucially, no bias toward any type of ubiquitin chain [10]. This unbiased recognition enables a more comprehensive capture of ubiquitinated proteins from complex biological samples, providing a more accurate representation of the cellular ubiquitination status.
Key Performance Metrics and Comparative Data
Rigorous comparative testing demonstrates the performance advantages of the ThUBD-coated platform across multiple critical parameters essential for reliable ubiquitination analysis.
Table 1: Quantitative Performance Comparison of ThUBD vs. TUBE Platforms
| Performance Parameter | ThUBD-Coated Plates | TUBE-Coated Plates | Improvement Factor |
|---|---|---|---|
| Detection Sensitivity | As low as 0.625 μg | ~10 μg | 16-fold |
| Dynamic Range | 16-fold wider linear range | Limited linear range | 16-fold wider |
| Ubiquitin Signal Enrichment | ~10-fold stronger signal | Baseline | 10-fold enhancement |
| Ubiquitin Chain Recognition | Unbiased toward all chain types | Bias toward specific linkages | Qualitative improvement |
| Assay Workflow | High-throughput, 96-well format | High-throughput, 96-well format | Equivalent |
The ThUBD-coated platform exhibits a remarkable 16-fold wider linear range for capturing polyubiquitinated proteins from complex proteome samples compared to TUBE-based methods [10]. This expanded dynamic range is crucial for accurately quantifying ubiquitination changes in response to experimental treatments, such as with PROTAC molecules.
Furthermore, when combined with specialized sample preparation techniques like Denatured-Refolded Ubiquitinated Sample Preparation (DRUSP), the ThUBD platform demonstrates the ability to yield a ubiquitin signal nearly three times stronger than conventional methods, with an overall enrichment efficiency approximately 10-fold higher than standard approaches [24]. This enhanced signal strength directly translates to improved detection of low-abundance ubiquitination events.
Experimental Protocols and Workflow Integration
ThUBD-Coated Plate Protocol for High-Throughput Ubiquitination Analysis
Coating Optimization: The foundational step involves coating 1.03 μg ± 0.002 of the ThUBD fusion protein onto Corning 3603-type 96-well plates, which have been empirically determined to provide optimal binding characteristics. This specific quantity enables the plate to bind approximately 5 pmol of polyubiquitin chains, maximizing the capture capacity without wasteful reagent usage [10].
Sample Processing: Cell lysates or complex proteome samples should be prepared using denaturing conditions to ensure complete protein extraction and to inhibit deubiquitinating enzymes (DUBs) and proteasome activity that would otherwise remove ubiquitin signals. The DRUSP method is particularly recommended, as it utilizes strongly denatured buffers for effective extraction, followed by a refolding step using filters to restore ubiquitin structure for recognition by ThUBD [24].
Enrichment and Detection: Add prepared samples to ThUBD-coated plates and incubate with gentle agitation. After thorough washing with optimized buffers to remove non-specifically bound proteins, captured ubiquitinated proteins can be detected using various methods including ThUBD-HRP conjugates for chemiluminescent detection, or eluted for downstream applications like Western blotting or mass spectrometry analysis [10].
Comparative Experimental Design for Method Validation
When validating these platforms for specific research applications, a direct comparative approach yields the most actionable data:
Parallel Processing: Split identical samples across both ThUBD-coated and TUBE-coated plates, following manufacturer protocols for each system. This controls for biological variability and enables direct performance comparison.
Dilution Series Analysis: Prepare serial dilutions of sample lysates to practically assess the sensitivity and dynamic range of each platform. This experimentally determines the minimum input requirement for reliable detection in a specific experimental context.
Specificity Controls: Include samples with known ubiquitination status (e.g., via overexpression of ubiquitin-GFP fusion constructs like Ub-GFP, Ub2-GFP, and Ub4-GFP) to verify the platform's ability to recognize different ubiquitin chain lengths and types [10].
Downstream Application: Elute captured proteins from both platforms and analyze them side-by-side using Western blotting with anti-ubiquitin antibodies or mass spectrometry to compare the diversity and abundance of ubiquitinated species enriched by each method.
Diagram 1: ThUBD-Coated Plate Workflow. This optimized protocol emphasizes denaturing conditions and precise coating parameters.
The Scientist's Toolkit: Essential Reagents and Materials
Successful implementation of high-throughput ubiquitination studies requires specific, quality-controlled reagents and materials. The following toolkit outlines essential components for establishing robust assays.
Table 2: Essential Research Reagent Solutions for UBD-Based Enrichment
| Reagent/Material | Function/Purpose | Specific Recommendations |
|---|---|---|
| ThUBD Fusion Protein | High-affinity, unbiased capture of ubiquitinated proteins | Recombinantly expressed and purified; 1.03 μg/well coating density [10] |
| 96-Well Plates | Solid support for assay | Corning 3603-type plates recommended for ThUBD coating [10] |
| Denaturing Lysis Buffer | Complete protein extraction, inhibits DUBs | Contains strong denaturants (e.g., SDS, urea) for DRUSP method [24] |
| Refolding Buffers | Restore ubiquitin structure after denaturation | Filter-based refolding system for DRUSP protocol [24] |
| Detection Reagents | Signal generation for quantification | ThUBD-HRP conjugate; chemiluminescent substrates [10] |
| Ubiquitin Standards | Assay validation and calibration | Ub-GFP, Ub2-GFP, Ub4-GFP fusion proteins [10] |
Contextualizing UBD-Based Enrichment in the Broader Research Landscape
Comparison with Antibody-Based Enrichment Methods
Ubiquitin-binding domains represent a distinct approach compared to traditional antibody-based enrichment. Antibodies against ubiquitin face inherent challenges due to the high conservation of ubiquitin itself, resulting in generally low-affinity reagents. Furthermore, commercial antibodies often exhibit significant bias toward specific ubiquitin chain types, potentially providing an incomplete picture of the ubiquitination landscape [10]. While antibody-coated surfaces have proven valuable for cell capture and enrichment in other applications [25], these limitations are particularly problematic for comprehensive ubiquitinome studies.
The UBD-based approach, particularly the ThUBD platform, addresses these fundamental limitations through engineered high-affinity interactions that recognize the structural features of ubiquitin and ubiquitin chains without preference for specific linkages. This technological difference translates to superior performance in research applications requiring a comprehensive view of ubiquitination status, such as in PROTAC development or pathway analysis [10].
Integration with High-Throughput Screening Paradigms
The microplate format of both ThUBD and TUBE technologies makes them inherently compatible with modern high-throughput screening (HTS) workflows. The standardized 96-well footprint ensures compatibility with automated liquid handlers, plate washers, and readers [26]. This compatibility is essential for applications in drug discovery, where profiling the effects of compound libraries on ubiquitination requires processing hundreds or thousands of samples in a reproducible, scalable manner.
The selection of appropriate microplates is a critical technical consideration often overlooked in assay development. Factors such as plate material, surface energy, well geometry, and binding capacity can significantly impact assay performance [26]. The recommendation for Corning 3603-type plates with ThUBD coating reflects empirical optimization to maximize signal-to-noise ratio and reproducibility in ubiquitination assays.
Diagram 2: Evolution of Ubiquitin Enrichment Technologies. The progression from antibodies to ThUBD shows improvements in affinity and reduction of linkage bias.
The comparative data presented in this guide demonstrates that ThUBD-coated plates offer significant advantages over TUBE-based methods for high-throughput ubiquitination studies. The 16-fold improvement in detection sensitivity and dynamic range, combined with unbiased recognition of all ubiquitin chain types, makes the ThUBD platform particularly suitable for applications requiring comprehensive ubiquitinome profiling or detection of subtle changes in ubiquitination status.
For research applications focused on specific, well-characterized ubiquitin linkages where maximum sensitivity is not critical, TUBE-based methods may still provide a cost-effective solution. However, for advanced applications in PROTAC development, biomarker discovery, and mechanistic studies of ubiquitination pathways, the ThUBD platform represents a superior tool that provides more accurate and comprehensive data. The optimized protocols and toolkit outlined in this guide provide researchers with a foundation for implementing these powerful enrichment technologies in their ubiquitination research workflows.
In mass spectrometry-based proteomics, sample preparation is a pivotal step that directly determines the depth, accuracy, and reliability of analytical results. This is especially true when investigating complex proteomes or working with low-input scenarios, where the starting biological material is limited. Efficient sample preparation concentrates substances of interest, removes impurities that cause background noise, and ensures results are both repeatable and reliable across different experiments [27]. The challenge intensifies in low-input proteomics (LIP), which deals with protein amounts in the mid- to low nanogram range, corresponding to approximately 1,000 mammalian cells or fewer, down to the single-cell level [28]. In these scenarios, traditional sample preparation methods often fail due to issues like analyte loss from surface adhesion and evaporation of small volume samples [28]. This guide focuses on evaluating two powerful enrichment strategies—UBD-based and antibody-based methods—within the specific context of ubiquitination research, providing researchers with a clear framework for selecting the optimal approach for their experimental needs.
Methodological Comparison: UBD-Based vs. Antibody-Based Enrichment
Fundamental Principles and Key Differentiators
UBD-Based Methods utilize engineered tandem hybrid ubiquitin-binding domains (ThUBDs) or similar constructs to capture ubiquitinated proteins. These domains are designed to recognize ubiquitin motifs with high affinity and without bias toward specific ubiquitin chain linkages [10]. The core advantage lies in their ability to provide unbiased recognition of all ubiquitin chain types, including mono-ubiquitination, multiple mono-ubiquitination, and various polyubiquitin chain linkages (K48, K63, K6, K11, K27, K29, K33, and M1-linked) [4] [10].
Antibody-Based Methods rely on immunocapture using anti-ubiquitin antibodies such as P4D1, FK1, or FK2 that recognize all ubiquitin linkages, or linkage-specific antibodies that target particular chain architectures [4]. These methods can be applied to endogenous proteins without genetic manipulation, making them suitable for clinical samples and animal tissues [4].
Table 1: Core Characteristics of UBD-Based and Antibody-Based Enrichment Methods
| Feature | UBD-Based Methods | Antibody-Based Methods |
|---|---|---|
| Basic Principle | Affinity capture via ubiquitin-binding domains | Immunoaffinity capture using ubiquitin antibodies |
| Linkage Recognition | Unbiased toward all ubiquitin chain types [10] | Pan-specific or linkage-specific antibodies available [4] |
| Genetic Modification | May require tagged Ub expression (for some approaches) [4] | Not required for endogenous protein studies [4] |
| Sample Compatibility | Cell lines, engineered systems [4] | Cell lines, animal tissues, clinical samples [4] |
| Key Advantage | High affinity, no linkage bias, suitable for high-throughput [10] | Can profile endogenous ubiquitination without genetic manipulation [4] |
Performance and Experimental Data Comparison
Recent technological advancements have yielded significant performance differences between these approaches. A high-throughput method using ThUBD-coated 96-well plates demonstrated a 16-fold wider linear range for capturing polyubiquitinated proteins from complex proteome samples compared to TUBE (Tandem Ubiquitin Binding Entity)-based plates [10]. The ThUBD-based platform showed significantly improved detection sensitivity, capable of capturing ubiquitinated proteins from sample amounts as low as 0.625 μg, substantially outperforming existing TUBE technology [10].
Antibody-based approaches, while powerful, face limitations including potential linkage bias with some antibodies, high cost, and non-specific binding that can reduce identification sensitivity [4]. However, they remain invaluable for studying endogenous ubiquitination in native systems, with linkage-specific antibodies enabling researchers to investigate the biology of specific ubiquitin chain types [4].
Table 2: Experimental Performance Metrics of Enrichment Methods
| Performance Metric | UBD-Based Methods | Antibody-Based Methods | Experimental Context |
|---|---|---|---|
| Detection Sensitivity | As low as 0.625 μg protein [10] | Not quantitatively specified in results | Complex proteome samples [10] |
| Dynamic Range | 16-fold wider than TUBE-based plates [10] | Not directly comparable | Linear range for capturing polyubiquitinated proteins [10] |
| Throughput Capacity | High-throughput 96-well plate format [10] | Lower throughput due to limitations in affinity reagents | Processing multiple samples simultaneously [10] |
| Identification Efficiency | Relatively low in tagging-based approaches [4] | Impairment due to non-specifically binding proteins [4] | Proteomic profiling of ubiquitinated substrates [4] |
Detailed Experimental Protocols
ThUBD-Based High-Throughput Enrichment Protocol
The ThUBD-coated plate method enables sensitive, high-throughput detection of ubiquitination signals. Below is the detailed experimental workflow:
Materials Required:
- Corning 3603-type 96-well plates [10]
- Recombinant ThUBD protein (1.03 μg per well for coating) [10]
- Binding/Wash Buffer: 50 mM Tris-HCl, 150 mM NaCl, 0.1% (v/v) Tween-20, pH 7.5 [10]
- Cell lysis buffer (e.g., containing protease inhibitors)
- ThUBD-HRP conjugate for detection [10]
- Appropriate chemiluminescent or colorimetric substrate
Procedure:
- Plate Coating: Coat each well of a 96-well plate with 1.03 μg of ThUBD protein in coating buffer. Incubate overnight at 4°C or for 2 hours at room temperature [10].
- Blocking: Block plates with a suitable blocking buffer (e.g., 3% BSA in PBS) for 1-2 hours at room temperature to prevent non-specific binding.
- Sample Preparation: Lyse cells in an appropriate lysis buffer. Centrifuge at 10,000-15,000 × g for 15 minutes to remove insoluble material. Determine protein concentration.
- Sample Incubation: Add diluted protein samples (as low as 0.625 μg total protein) to ThUBD-coated wells. Incubate for 2 hours at room temperature with gentle shaking [10].
- Washing: Wash wells 3-5 times with Binding/Wash Buffer to remove unbound proteins and contaminants [10].
- Detection: Incubate with ThUBD-HRP conjugate for 1 hour at room temperature. After washing, add appropriate substrate and measure signal using a plate reader [10].
Optimization Notes: The Corning 3603 plate has been identified as optimal for ThUBD coating. Coating conditions, washing buffer composition, and detection conditions should be systematically optimized for specific experimental setups [10].
Antibody-Based Ubiquitin Enrichment Protocol
Materials Required:
- Anti-ubiquitin antibodies (e.g., P4D1, FK1, FK2 for pan-specific detection or linkage-specific antibodies) [4]
- Protein A/G agarose or magnetic beads
- Cell lysis buffer (e.g., RIPA buffer with protease inhibitors)
- Wash buffers of varying stringency (e.g., with increasing salt concentrations)
- Elution buffer (e.g., low pH buffer or SDS-PAGE sample buffer)
Procedure:
- Sample Preparation: Lyse cells in an appropriate lysis buffer. Centrifuge to remove insoluble material. Determine protein concentration.
- Antibody-Bead Preparation: Incubate anti-ubiquitin antibody with Protein A/G beads for 1-2 hours at 4°C to form antibody-bead complexes.
- Immunoprecipitation: Incubate protein lysate with antibody-bead complexes overnight at 4°C with gentle rotation.
- Washing: Pellet beads and wash 3-5 times with wash buffers of increasing stringency to remove non-specifically bound proteins.
- Elution: Elute bound ubiquitinated proteins using low pH buffer (e.g., 0.1 M glycine, pH 2.5-3.0) or by boiling in SDS-PAGE sample buffer.
- Downstream Analysis: Process eluted proteins for Western blotting or mass spectrometry analysis.
Research Reagent Solutions Toolkit
Table 3: Essential Reagents for Ubiquitin Enrichment Studies
| Reagent/Category | Specific Examples | Function & Application |
|---|---|---|
| Affinity Tags | 6× His tag, Strep-tag [4] | Purification of ubiquitinated substrates; enables screening in cells [4] |
| Ubiquitin Antibodies | P4D1, FK1/FK2 (pan-specific); linkage-specific antibodies [4] | Enrich and detect ubiquitinated substrates; study specific chain linkages [4] |
| UBD Reagents | TUBE, ThUBD [10] | High-affinity capture of ubiquitinated proteins with minimal linkage bias [10] |
| Cell Lysis Reagents | Urea, SDC, Protease inhibitors [29] | Efficient extraction while preserving protein modifications and activity |
| Enrichment Supports | Ni-NTA agarose (His tag), Strep-Tactin (Strep tag) [4] | Resins for affinity purification of tagged ubiquitinated proteins [4] |
| Detection Reagents | ThUBD-HRP, Chemiluminescent substrates [10] | Signal generation for quantification of captured ubiquitinated proteins [10] |
Workflow Visualization
The diagram above illustrates the parallel pathways for UBD-based and antibody-based enrichment methods, highlighting their convergence at the washing and elution steps before final analysis. This visualization underscores the shared technical requirements but different affinity principles of each approach.
The systematic comparison between UBD-based and antibody-based enrichment methods reveals a clear trade-off between analytical performance and practical applicability. UBD-based methods, particularly those employing advanced ThUBD technology, offer superior sensitivity, dynamic range, and throughput capacity, making them ideal for high-throughput screening applications in drug discovery, particularly in PROTAC development [10]. The 16-fold improvement in detection range and ability to work with sub-microgram protein amounts positions ThUBD-based platforms as the preferred choice for quantitative studies requiring high sensitivity [10].
Conversely, antibody-based methods maintain their importance for studies of endogenous ubiquitination in clinical samples and animal tissues where genetic manipulation is infeasible [4]. The availability of linkage-specific antibodies further enables targeted investigation of specific ubiquitin chain biology, though researchers must remain cognizant of potential linkage bias and higher costs associated with this approach [4].
For researchers handling complex proteomes and low-input scenarios, the selection between these methods should be guided by specific experimental needs: UBD-based methods for maximum sensitivity and throughput with engineered systems, and antibody-based approaches for endogenous ubiquitination studies in native biological samples. As both technologies continue to evolve, their complementary strengths will undoubtedly provide increasingly powerful tools for deciphering the complex landscape of protein ubiquitination in health and disease.
Ubiquitination is a fundamental post-translational modification that regulates virtually all aspects of eukaryotic cell biology, governing processes from protein degradation to DNA repair and immune signaling [9] [4]. The remarkable diversity of ubiquitin signaling stems from the ability of ubiquitin to form polymeric chains through different linkage types between its amino acid residues, creating a complex "ubiquitin code" that determines specific cellular outcomes [9]. Currently, researchers face a methodological crossroads when designing studies to decipher this code: whether to employ linkage-specific antibodies or broad-spectrum ubiquitin-binding domains (UBDs) for the enrichment and detection of ubiquitinated substrates. This guide provides an objective comparison of these approaches, supported by experimental data and methodological considerations, to inform researchers' experimental design decisions.
The ubiquitin system's complexity is staggering—with more than 110,000 ubiquitination sites identified in human cells across over 12,000 proteins [9]. Ubiquitin can be attached to substrates as single monomers or as polymers of eight canonical amide linkages (M1, K6, K11, K27, K29, K33, K48, K63) and four recently discovered ester linkages (T12, T14, S20, T22) [9]. Each linkage type adopts a distinct three-dimensional structure that enables specific functions, with K48-linked chains typically targeting substrates for proteasomal degradation and K63-linked chains mediating non-degradative signaling in pathways like DNA damage response and immune signaling [9] [4]. This linkage specificity forms the foundation of ubiquitin's signaling versatility, making the development of precise detection tools paramount to advancing our understanding of cellular regulation.
Technical Comparison: Mechanisms and Methodologies
Molecular Recognition Mechanisms
Linkage-specific antibodies are immunoglobulin-based reagents engineered to recognize unique structural epitopes presented by specific polyubiquitin chain linkages. These antibodies typically target the characteristic isopeptide bond regions between ubiquitin monomers, which display distinct conformations based on the connected lysine or methionine residue [9]. For example, antibodies specific for K48-linked chains recognize the compact, closed structure formed by this linkage type, while those for K63-linked chains target the more extended, open conformation [4]. The manufacturing process involves immunizing animals with synthetically generated di-ubiquitin of defined linkage or through phage display screening to obtain high-affinity binders that can discriminate between structurally similar linkages.
Broad-spectrum UBDs encompass a diverse family of natural protein domains that recognize common surfaces on ubiquitin, primarily the hydrophobic "Ile44 patch" present on all ubiquitin molecules regardless of linkage type [30]. These domains include NZF (Npl4 zinc finger), UBA (ubiquitin-associated), UIM (ubiquitin-interacting motif), and others that exhibit varying degrees of linkage preference through auxiliary interaction surfaces [30]. Unlike antibodies, UBDs are typically smaller (30-50 amino acids for NZF domains) and can be engineered as tandem repeats to enhance affinity through avidity effects [4] [30]. Recent research has revealed that some NZF domains achieve surprising specificity through secondary interaction sites that contact both ubiquitin and the modified substrate simultaneously [30].
Table 1: Key Research Reagent Solutions for Ubiquitin Enrichment
| Reagent Type | Specific Examples | Key Features | Primary Applications |
|---|---|---|---|
| Linkage-Specific Antibodies | Anti-K48, Anti-K63, Anti-M1 (linear) | High linkage specificity, commercial availability, recognize defined epitopes | Immunoblotting, immunofluorescence, immunohistochemistry, enrichment for MS |
| Broad-Spectrum UBDs | Tandem UBA domains, NZF arrays, engineered UBQs | Recognize common ubiquitin surfaces, modular design, can be mutagenized | Affinity purification, proteomic screening, in vitro binding assays |
| Ubiquitin Tags | His-Ub, Strep-Ub, HA-Ub | Genetic encoding, affinity handle incorporation, whole-protein purification | Identification of ubiquitinated substrates, ubiquitination site mapping |
| General Ubiquitin Antibodies | P4D1, FK1, FK2 | Pan-ubiquitin recognition, detect various linkage types | General ubiquitination detection, immunoblotting of total ubiquitin |
Experimental Workflows and Protocols
The fundamental workflows for both linkage-specific antibodies and UBDs share common steps but differ in critical aspects that influence experimental outcomes. The diagram below illustrates the core methodological pathways for each approach:
Standard Protocol for Linkage-Specific Antibody Enrichment:
- Antibody Immobilization: Covalently cross-link 2-5 µg of linkage-specific antibody to 20 µL of protein A/G beads using DSS or similar cross-linker (30-60 minute incubation).
- Sample Preparation: Lyse cells in RIPA buffer supplemented with 1% SDS, followed by 5-10× dilution to reduce SDS concentration to 0.1-0.2%. Include N-ethylmaleimide (5-10 mM) to inhibit deubiquitinases and protease/phosphatase inhibitors.
- Immunoprecipitation: Incubate 500-2000 µg of protein lysate with immobilized antibodies for 2-4 hours at 4°C with gentle rotation.
- Washing: Perform 3-5 washes with modified RIPA buffer (150 mM NaCl, 0.1% SDS) to reduce non-specific binding.
- Elution: Elute bound proteins using 2× Laemmli buffer at 95°C for 10 minutes or low-pH glycine buffer (pH 2.5-3.0) followed by neutralization.
- Analysis: Process eluates for western blotting or mass spectrometry analysis.
Standard Protocol for Broad-Spectrum UBD Enrichment:
- UBD Immobilization: Incubate 10-20 µg of GST- or His-tagged UBD construct with appropriate affinity resin (glutathione or nickel beads) for 30-60 minutes.
- Sample Preparation: Lyse cells in non-denaturing buffer (e.g., NP-40 or Triton X-100-based) to preserve protein interactions. Include deubiquitinase inhibitors.
- Binding Reaction: Incubate 500-2000 µg of protein lysate with immobilized UBDs for 1-2 hours at 4°C.
- Washing: Perform 2-3 washes with lysis buffer containing 150-300 mM NaCl.
- Elution: Compete off bound proteins using free ubiquitin (1-2 mg/mL) or cleave with PreScission protease if using GST-tagged constructs with cleavage sites.
- Analysis: Process eluates for downstream applications.
Performance Comparison: Sensitivity, Specificity, and Applications
Quantitative Performance Metrics
Table 2: Direct Performance Comparison of Enrichment Methods
| Performance Metric | Linkage-Specific Antibodies | Broad-Spectrum UBDs | Experimental Basis |
|---|---|---|---|
| Linkage Specificity | High for target linkage (K48/K63: >95%) | Variable (linkage preference vs. broad recognition) | Specific antibodies show minimal cross-reactivity; UBDs show graded preferences [4] [30] |
| Affinity Range | High (nM-pM Kd values) | Moderate to low (μM-mM Kd values) | Antibodies engineered for high affinity; natural UBDs have weaker monomeric affinity [30] |
| Enrichment Efficiency | 50-80% for target linkage | 30-60% for total ubiquitin | Comparative proteomic studies showing differential recovery [4] [31] |
| Background Binding | Low with optimized washing | Moderate to high | Antibodies show cleaner backgrounds in blotting applications [4] |
| Multiplexing Capacity | Limited by antibody availability | High with different UBD types | UBD panels can enrich broader ubiquitin landscapes [9] |
| Compatibility with Denaturing Conditions | Good (0.1-1% SDS) | Poor (structure-dependent binding) | Antibodies maintain recognition under mild denaturation [4] |
Experimental Evidence and Case Studies
Recent studies provide compelling comparative data on the performance of these methodologies. In CAR NK cell detection research, linker-specific monoclonal antibodies demonstrated exceptional specificity in detecting engineered immunoreceptors, showing no cross-reactivity with CARs containing alternative linker sequences [32]. This high specificity enabled clear distinction between positive and negative populations in flow cytometry, outperforming polyclonal detection methods which showed significant fluctuation and dim signals [32].
In ubiquitin research, comprehensive analysis of NZF UBD domains revealed that while most NZF domains do not display strong chain linkage preference, they achieve remarkable substrate specificity through secondary interaction surfaces that simultaneously recognize both ubiquitin and the modified substrate [30]. For instance, the TAB2 NZF domain preferentially binds phosphorylated K6 chains present at depolarized mitochondria, while the HOIP NZF1 domain specifically recognizes monoubiquitinated forms of NEMO and optineurin [30]. This demonstrates how UBDs can provide contextual specificity beyond simple linkage recognition.
The UbiCRest method (a UBD-based approach) has been systematically compared with antibody-based methods for ubiquitin chain architecture analysis, revealing complementary strengths. While linkage-specific antibodies provide unambiguous identification of particular chain types, UBD-based approaches can reveal mixed and branched chains that might be missed by single-specificity antibodies [31].
Decision Framework: Selecting the Appropriate Method
Application-Specific Recommendations
The choice between linkage-specific antibodies and broad-spectrum UBDs should be guided by the specific research question and experimental context. The following decision framework illustrates key considerations:
Scenarios Favoring Linkage-Specific Antibodies:
- Hypothesis-Driven Research: When investigating the role of a specific ubiquitin linkage type with known cellular functions (e.g., K48-mediated degradation or K63-mediated signaling).
- Quantitative Applications: When precise quantification of a particular chain type is required, as in diagnostic assays or therapeutic monitoring.
- Spatial Localization Studies: For immunohistochemistry or immunofluorescence where antibody specificity enables subcellular localization of specific ubiquitin linkages.
- Clinical Samples: When working with patient tissues where genetic manipulation for tagged ubiquitin expression is impossible [4].
Scenarios Favoring Broad-Spectrum UBDs:
- Discovery Proteomics: When conducting unbiased screens for ubiquitinated substrates across multiple linkage types.
- Native Interaction Studies: When preserving protein complexes and interaction networks is essential for understanding biological context.
- Structural Biology Applications: When the UBD itself is part of the mechanistic study or when engineering customized binding domains.
- Investigating Atypical Linkages: When studying less common ubiquitin linkages where high-quality antibodies may be unavailable [30].
Integrated Approaches and Future Directions
Increasingly, sophisticated research questions require integrated methodologies that leverage the strengths of both approaches. Sequential enrichment strategies—using broad-spectrum UBDs for initial capture followed by linkage-specific antibodies for refinement—can provide unprecedented resolution of the ubiquitin landscape [4] [31]. Additionally, new engineered systems like the "Ubiquiton" platform, which enables inducible, linkage-specific polyubiquitylation of target proteins, provide validation tools that complement both antibody and UBD-based detection methods [33].
Emerging technologies are also bridging the gap between these approaches. For instance, bead-based biosensing platforms that couple the specificity of antibodies with the signal amplification of UBD-based detection systems show promise for highly sensitive diagnostic applications [34] [35]. Similarly, advances in mass spectrometry instrumentation and sample preparation are enhancing the compatibility of both enrichment methods with proteomic analysis, enabling more comprehensive ubiquitinome mapping [4] [31].
Table 3: Troubleshooting Guide for Common Experimental Challenges
| Challenge | Antibody-Based Solutions | UBD-Based Solutions |
|---|---|---|
| High Background | Increase salt concentration in washes (150-300 mM NaCl); include mild detergents; use cross-linked antibodies | Optimize UBD concentration; include competitor (BSA or irrelevant protein); use tandem UBDs for cleaner enrichment |
| Low Yield | Increase antibody amount; extend incubation time; optimize lysis conditions | Use higher-affinity UBD constructs; engineer tandem domains; reduce salt concentration in binding buffer |
| Loss of Specificity | Titrate antibody amount; include specific competing antigen; verify linkage specificity with controls | Utilize UBD mutants with narrowed specificity; employ binding conditions that favor specific interactions |
| Incompatibility with Downstream Applications | Switch elution method (low pH vs. boiling); buffer exchange after elution; test multiple extraction protocols | Use cleavable tags for gentle elution; optimize buffer conditions for specific analytical platforms |
The decision between linkage-specific antibodies and broad-spectrum UBDs represents a fundamental methodological choice in ubiquitin research that significantly influences experimental outcomes. Linkage-specific antibodies offer superior specificity for defined ubiquitin chain types and perform well under diverse experimental conditions, making them ideal for targeted studies and diagnostic applications. Broad-spectrum UBDs provide more versatile tools for discovery proteomics and native interaction studies, with the potential for engineering enhanced specificity through multivalent interactions. The most sophisticated approaches increasingly combine both methodologies in sequential or parallel strategies to leverage their complementary strengths. As the ubiquitin field continues to evolve, methodological innovations will further refine these essential tools, enabling researchers to progressively decipher the complex language of the ubiquitin code with greater precision and biological relevance.
The identification and analysis of low-abundance proteins and specific post-translational modifications (PTMs) represent a significant challenge in modern proteomics. Protein ubiquitination, a crucial PTM that regulates diverse cellular functions including protein degradation, cell signaling, and DNA repair, is particularly difficult to study due to its low stoichiometry, transient nature, and complexity of ubiquitin chain architectures [4]. To overcome these challenges, effective enrichment techniques are essential as a bridge to downstream analysis methods such as mass spectrometry (MS) and immunoblotting. Without robust enrichment strategies, the target analytes remain obscured by high-abundance proteins, rendering them virtually undetectable by even the most sensitive analytical platforms [36].
This guide focuses on evaluating two primary enrichment approaches: ubiquitin-binding domain (UBD)-based methods and antibody-based techniques. We will objectively compare their performance characteristics, provide supporting experimental data, and detail methodologies to help researchers select the optimal strategy for their specific applications in drug development and basic research.
Enrichment Methodologies: Principles and Technical Considerations
Ubiquitin-Binding Domain (UBD)-Based Enrichment
UBD-based enrichment utilizes engineered protein domains that specifically recognize and bind to ubiquitin moieties attached to substrate proteins. The fundamental principle involves exploiting the natural affinity between UBDs and ubiquitin molecules, often enhanced through protein engineering to improve binding characteristics [10] [37]. These domains can be designed to capture all ubiquitinated proteins indiscriminately or to selectively enrich for specific ubiquitin chain linkages, depending on the research objectives.
Recent advances in UBD technology have led to the development of Tandem Hybrid Ubiquitin Binding Domains (ThUBDs), which combine multiple ubiquitin-recognition elements to achieve higher affinity and broader specificity compared to single UBDs [10]. ThUBDs exhibit unbiased recognition of different ubiquitin chain types while demonstrating significantly improved binding capacity for polyubiquitinated proteins from complex proteome samples [10]. The binding process relies on the proper spatial structure of ubiquitin and ubiquitin chains, requiring non-denaturing conditions to preserve these structural interactions during the enrichment process [37].
Antibody-Based Enrichment
Antibody-based enrichment employs immunoglobulin molecules raised against ubiquitin or specific ubiquitin chain linkages to immunoprecipitate ubiquitinated proteins from complex mixtures. This approach leverages the high specificity of antigen-antibody interactions, with antibodies typically targeting the ubiquitin protein itself or distinctive epitopes formed by specific ubiquitin chain linkages [4].
Common antibodies used in ubiquitination studies include P4D1, FK1, and FK2, which recognize various forms of ubiquitin, alongside linkage-specific antibodies targeting M1-, K11-, K27-, K48-, or K63-linked chains [4]. The antibody-based approach allows for the study of endogenous ubiquitination under physiological conditions without genetic manipulation of the target system, making it particularly valuable for clinical samples and animal tissues where genetic tagging is infeasible [4]. However, challenges include the high cost of high-quality antibodies, potential linkage bias, and limited availability of antibodies capable of recognizing all ubiquitin chain types equally well [4].
Comparative Performance Analysis: UBD-Based vs. Antibody-Based Methods
Quantitative Performance Metrics
Table 1: Direct comparison of key performance metrics between UBD-based and antibody-based enrichment methods
| Performance Metric | UBD-Based Methods | Antibody-Based Methods | Experimental Context |
|---|---|---|---|
| Detection Sensitivity | 16-fold wider linear range vs. TUBE [10] | Not explicitly quantified | Capture of polyubiquitinated proteins from complex proteomes [10] |
| Dynamic Range | As low as 0.625 μg [10] | Limited by antibody affinity | Amount of input material required for detection [10] |
| Linkage Recognition | Unbiased recognition of all ubiquitin chain types [10] | Bias toward specific ubiquitin chains [4] | Capacity to detect different ubiquitin chain linkages [4] [10] |
| Signal Intensity | ~10-fold stronger ubiquitin signal vs. control [37] | Variable depending on antibody quality | Ubiquitin signal enrichment efficiency [37] |
| Sample Compatibility | Requires native conditions or refolding [37] | Compatible with various conditions including denatured | Flexibility with sample preparation protocols [4] |
| Throughput Capacity | High-throughput 96-well plate format [10] | Typically lower throughput | Adaptation to automated screening platforms [10] |
Experimental Workflows and Methodologies
ThUBD-Coated Plate Methodology for High-Throughput Ubiquitination Analysis
The ThUBD-coated plate technology represents a significant advancement in UBD-based enrichment, enabling high-throughput screening of ubiquitination events. The experimental protocol involves several key steps:
Plate Preparation and Coating: Corning 3603-type 96-well plates are coated with 1.03 μg ± 0.002 of ThUBD fusion protein, which has been previously expressed and purified from recombinant E. coli strains. The coating process is optimized to ensure uniform distribution and maximal binding capacity, with each well capable of specifically binding approximately 5 pmol of polyubiquitin chains [10].
Sample Preparation and Binding: Complex proteome samples are extracted under native conditions to preserve ubiquitin structure, with protein concentrations carefully determined. Between 0.625 μg to 10 μg of total protein lysate is added to each well and incubated for 2 hours at 4°C with gentle agitation to facilitate specific binding between ThUBD and ubiquitinated proteins [10].
Washing and Elution: Non-specifically bound proteins are removed through multiple washing steps with optimized wash buffers that maintain protein stability while eliminating contaminants. Bound ubiquitinated proteins are then eluted using conditions that disrupt the ThUBD-ubiquitin interaction without denaturing the proteins of interest [10].
Downstream Analysis: The eluted proteins can be directly analyzed by Western blotting using specific antibodies, or prepared for mass spectrometry analysis through digestion and peptide purification [10].
This methodology demonstrates particular strength in PROTAC drug development applications, where monitoring dynamic changes in protein ubiquitination is essential for evaluating compound efficacy [10].
Denatured-Refolded Ubiquitinated Sample Preparation (DRUSP) Coupled with ThUBD
A recent innovative approach addresses the limitation of UBD-based methods that typically require native conditions. The DRUSP methodology combines strong denaturation with a refolding step to enhance ubiquitinated protein recovery:
Denaturing Extraction: Samples are lysed using strongly denatured buffers (e.g., containing high concentrations of urea or SDS) to ensure complete protein extraction, including insoluble fractions, while simultaneously inactivating deubiquitinating enzymes (DUBs) and proteasomes that would otherwise remove ubiquitin signals [37].
Filter-Based Refolding: The denatured extracts are subjected to a refolding process using specialized filters that gradually remove denaturants, allowing ubiquitin and ubiquitin chains to regain their native spatial structures necessary for recognition by ThUBD or other UBDs [37].
Enrichment and Analysis: The refolded samples are then applied to ThUBD-based enrichment platforms following standard protocols, with subsequent analysis by MS or immunoblotting [37].
This hybrid approach demonstrates a significant improvement in ubiquitin signal recovery, with approximately 10-fold higher enrichment efficiency compared to conventional methods under native conditions alone [37].
Antibody-Based Immunoprecipitation Protocol
Traditional antibody-based enrichment follows a well-established protocol:
Antibody Immobilization: Specific anti-ubiquitin antibodies (e.g., FK2 for total ubiquitin or linkage-specific antibodies) are coupled to protein A/G agarose beads or magnetic beads through incubation for 2-4 hours at 4°C [4] [38].
Immunoprecipitation: Cell or tissue lysates prepared under non-denaturing or mildly denaturing conditions are incubated with the antibody-bead complexes overnight at 4°C to allow specific binding [4].
Washing and Elution: Beads are extensively washed with appropriate buffers to remove non-specifically bound proteins, followed by elution of the ubiquitinated proteins using Laemmli buffer for Western blotting or specific elution conditions for mass spectrometry analysis [4] [38].
This approach has been successfully used in various studies, including the identification of 96 ubiquitination sites from human MCF-7 breast cancer cells using FK2 affinity chromatography [4].
Applications in Drug Development
The evaluation of enrichment efficiency has particular significance in drug development, especially in the emerging field of Proteolysis-Targeting Chimeras (PROTACs). Both UBD-based and antibody-based methods provide critical tools for:
Target Engagement Assessment: Confirming that PROTAC molecules successfully induce ubiquitination of intended target proteins [10].
Degradation Efficiency Quantification: Measuring the correlation between ubiquitination levels and subsequent protein degradation [10].
Mechanistic Studies: Elucidating the specificity of E3 ligase recruitment and ubiquitin chain topology formation [10] [37].
High-throughput UBD-based platforms offer distinct advantages for screening applications in pharmaceutical development, where rapid assessment of multiple candidates is essential [10].
Integrated Workflow for Downstream Analysis
Diagram 1: Integrated downstream analysis workflow bridging enrichment methods to analytical platforms. Both UBD-based and antibody-based enrichment techniques serve as critical preparation steps for mass spectrometry and immunoblotting analysis, enabling comprehensive characterization of ubiquitinated proteins.
Research Reagent Solutions for Ubiquitin Enrichment
Table 2: Essential research reagents and materials for ubiquitination studies
| Reagent/Material | Function | Application Examples |
|---|---|---|
| ThUBD-coated plates | High-affinity, unbiased capture of ubiquitinated proteins in high-throughput format | Global ubiquitination profiling, PROTAC screening [10] |
| TUBE (Tandem Ubiquitin Binding Entities) | General capture of polyubiquitinated proteins under native conditions | Conventional ubiquitin enrichment, interaction studies [37] |
| Linkage-specific UBDs | Selective enrichment of specific ubiquitin chain types | Studying particular ubiquitin signaling pathways [37] |
| Anti-ubiquitin antibodies (P4D1, FK1/FK2) | Immunoprecipitation of total ubiquitinated proteins | Target-specific ubiquitination analysis, clinical samples [4] |
| Linkage-specific ubiquitin antibodies | Selective detection of specific ubiquitin chain linkages | Studying chain-specific ubiquitination events [4] |
| DRUSP (Denatured-Refolded Ubiquitinated Sample Preparation) | Enhanced extraction and refolding for improved ubiquitin signal recovery | Difficult samples (fibrotic tissues, aggregates), improved reproducibility [37] |
| His-tagged Ubiquitin | Affinity purification of ubiquitinated proteins via metal chromatography | Ubiquitinome studies in genetically modifiable systems [4] |
| Strep-tagged Ubiquitin | Affinity purification using Strep-Tactin resin | Alternative tagging approach for ubiquitin pull-down [4] |
The selection between UBD-based and antibody-based enrichment methods represents a critical decision point in ubiquitination studies that significantly impacts downstream analytical outcomes. UBD-based methods, particularly recent advances like ThUBD platforms and DRUSP methodologies, offer superior performance in terms of sensitivity, dynamic range, and linkage unbiasedness, making them ideal for discovery-phase research and high-throughput drug screening applications. Antibody-based methods maintain utility for targeted studies, particularly when working with clinical specimens or when specific chain linkages are of interest.
The integration of robust enrichment protocols with sophisticated downstream analysis platforms remains essential for advancing our understanding of ubiquitination biology and accelerating the development of novel therapeutics targeting the ubiquitin-proteasome system. Researchers should carefully consider their specific experimental requirements, sample availability, and analytical goals when selecting the most appropriate enrichment strategy for their ubiquitination studies.
Solving Common Pitfalls and Enhancing Enrichment Performance
Overcoming Low Affinity and Linkage Bias in Antibody-Based Methods
Protein ubiquitination, a crucial post-translational modification, regulates diverse cellular functions including protein degradation, DNA repair, and immune responses. Research in this field relies heavily on the ability to accurately detect and characterize ubiquitinated proteins and their complex chain architectures. Currently, two primary methodological approaches dominate the field: antibody-based methods and ubiquitin-binding domain (UBD)-based methods. Antibody-based approaches have traditionally utilized antibodies that recognize ubiquitin or specific ubiquitin chain linkages, while UBD-based methods employ natural protein domains that inherently bind ubiquitin. Both approaches face significant technical challenges related to binding affinity and linkage bias, which can substantially impact their enrichment efficiency and the accuracy of experimental results. Affinity issues are particularly problematic when studying low-abundance ubiquitination events or when working with limited biological samples, potentially leading to false negatives. Linkage bias presents an even more insidious problem, as it can skew research findings toward well-characterized ubiquitin chain types while neglecting less common but biologically important linkages. This methodological comparison guide provides an objective evaluation of current technologies, focusing specifically on their capabilities to overcome these fundamental limitations, with particular attention to quantitative performance metrics and experimental validation data that directly inform researcher selection of appropriate methods for specific ubiquitination studies.
Methodological Approaches and Their Limitations
Antibody-Based Methods: Specificity at a Cost
Antibody-based methods represent the traditional workhorse for ubiquitination detection, leveraging the specificity of antibodies raised against ubiquitin or specific ubiquitin chain linkages. These approaches typically utilize immunoblotting techniques with anti-ubiquitin antibodies such as P4D1, FK1, and FK2, which recognize all ubiquitin linkages, or linkage-specific antibodies targeting M1-, K11-, K27-, K48-, or K63-linked chains [4]. The primary advantage of antibody-based methods lies in their ability to detect endogenous ubiquitinated proteins without requiring genetic manipulation, making them applicable to clinical samples and animal tissues [4] [31]. Additionally, linkage-specific antibodies enable researchers to investigate the biological functions associated with particular ubiquitin chain types, such as the well-established role of K48-linked chains in targeting substrates for proteasomal degradation [4].
However, antibody-based methods suffer from significant limitations that can compromise their effectiveness in ubiquitination research. A major constraint is the relatively low affinity of many commercially available anti-ubiquitin antibodies, which struggle to efficiently capture ubiquitinated proteins from complex proteomic samples, particularly when studying low-abundance targets or limited sample quantities [10]. This affinity limitation directly impacts detection sensitivity and can result in false negatives. Perhaps more problematic is the issue of linkage bias, where antibodies exhibit preferential recognition of certain ubiquitin chain types over others [10]. This bias creates substantial blind spots in ubiquitination profiling and can lead to misleading biological interpretations. Additional practical constraints include the high cost of high-quality antibodies and considerable batch-to-batch variability, which affects experimental reproducibility [4] [31]. These limitations collectively restrict the utility of antibody-based methods for comprehensive ubiquitination profiling, particularly when unbiased detection of diverse ubiquitin chain architectures is required.
UBD-Based Methods: Toward Unbiased Detection
UBD-based methods offer a complementary approach to antibody-based detection by utilizing natural ubiquitin-binding domains present in various cellular proteins. These domains, which include UIM, UBA, and NZF domains among others, inherently recognize and bind to ubiquitin moieties [4]. Early UBD-based approaches typically employed single UBDs for ubiquitin enrichment, but these suffered from low affinity that limited their practical utility [4]. Technological advancements have addressed this limitation through the development of tandem UBD arrangements, such as Tandem Ubiquitin-Binding Entities (TUBEs), which significantly improve affinity through avidity effects [10]. The most significant advantage of UBD-based methods is their potential for reduced linkage bias compared to antibody-based approaches, as certain UBD designs can recognize a broader spectrum of ubiquitin chain architectures [10].
Despite these advantages, conventional UBD-based methods still face important challenges. While improved over single UBDs, the binding affinity of first-generation TUBEs remains suboptimal for efficiently capturing polyubiquitinated proteins from complex biological samples [10]. Similar to antibody-based methods, some UBDs still exhibit preferential binding to specific ubiquitin chain types, creating persistent linkage bias concerns [4]. Additionally, UBD-based methods generally demonstrate lower efficiency for capturing monoubiquitinated proteins compared to polyubiquitinated species, creating a detection gap for important monoubiquitination events [31]. There is also potential for non-specific binding of other cellular proteins to the UBDs or adjacent purification tags, increasing background noise and reducing specificity [31]. These limitations have prompted the development of next-generation UBD technologies designed to overcome these persistent challenges.
Comparative Performance Analysis
The following tables provide a systematic comparison of the key characteristics and quantitative performance metrics for different ubiquitination detection methods, with particular focus on recently developed technologies that address affinity and linkage bias limitations.
Table 1: Comparison of Ubiquitination Enrichment Method Characteristics
| Method | Affinity | Linkage Recognition | Key Advantages | Major Limitations |
|---|---|---|---|---|
| Antibody-Based | Variable; often low | Typically linkage-specific (biased) | Works with endogenous proteins; applicable to clinical samples | High cost; batch variability; limited sensitivity |
| TUBE-Based | Moderate | Reduced bias vs. antibodies | Can protect ubiquitinated proteins from deubiquitinases | Still exhibits some linkage preference; moderate affinity |
| ThUBD-Based | High (Kd ~ nM range) | Unbiased recognition of all major chain types | High sensitivity and wide dynamic range | Requires recombinant protein production |
Table 2: Quantitative Performance Comparison of Detection Platforms
| Performance Metric | TUBE-Coated Plates | ThUBD-Coated Plates | Improvement Factor |
|---|---|---|---|
| Detection Sensitivity | ~10 μg | 0.625 μg | 16-fold |
| Linear Dynamic Range | Narrow | Wide (0.625-80 μg) | 16-fold wider |
| PolyUb Chain Capture | Moderate efficiency | High efficiency (~5 pmol capacity) | Significantly improved |
| Linkage Coverage | Partial bias | Unbiased for major chain types | Substantially more comprehensive |
The performance data clearly demonstrates the substantial advantages of the ThUBD-coated platform over conventional TUBE-based approaches [10]. Most notably, the ThUBD technology achieves a 16-fold improvement in detection sensitivity, enabling reliable detection of ubiquitinated proteins at quantities as low as 0.625 μg compared to the 10 μg required by TUBE-based methods [10]. This enhanced sensitivity is particularly valuable when working with precious or limited biological samples. Additionally, the ThUBD platform provides a 16-fold wider linear dynamic range (0.625-80 μg), allowing for more accurate quantification of ubiquitination levels across diverse sample types and concentrations [10]. Perhaps most importantly for comprehensive ubiquitination studies, the ThUBD technology exhibits unbiased recognition of all major ubiquitin chain types, addressing a fundamental limitation of both antibody-based and conventional UBD-based methods [10].
Experimental Protocols and Validation
ThUBD-Coated Plate Methodology
The development and validation of the ThUBD-coated plate platform followed a rigorous experimental workflow designed to maximize performance while ensuring reproducibility. The process began with the expression and purification of the Tandem Hybrid Ubiquitin-Binding Domain (ThUBD) fusion protein, which was previously engineered in the laboratory to combine favorable binding properties from different natural UBDs [10]. Researchers systematically screened various high-capacity 96-well plates to identify the optimal solid support, ultimately selecting Corning 3603-type plates based on their superior binding characteristics [10]. Critical coating conditions were optimized through iterative testing, establishing that coating with 1.03 μg ± 0.002 of ThUBD per well provided optimal performance while maintaining consistency across plates [10]. The washing buffer composition and detection conditions were similarly refined to minimize non-specific background while maximizing signal-to-noise ratios for ubiquitinated protein detection [10].
Validation experiments demonstrated that the optimized ThUBD-coated plates could specifically bind approximately 5 pmol of polyubiquitin chains under standard assay conditions [10]. The platform was shown to efficiently capture ubiquitinated proteins modified with diverse chain types, including K48-, K63-, and M1-linked chains, confirming the unbiased binding characteristics of the ThUBD technology [10]. Importantly, the coated plates maintained consistent performance over extended storage periods and across different production batches, addressing the reproducibility concerns commonly associated with antibody-based methods [10].
Comparative Experimental Workflow
The experimental workflow for evaluating ubiquitination detection methods involved a side-by-side comparison of ThUBD-coated plates, TUBE-coated plates, and antibody-based approaches using standardized samples and conditions. The process began with sample preparation, including complex proteome samples from HEK293T cells and purified ubiquitinated proteins (GFP, Ub-GFP, Ub2-GFP, and Ub4-GFP) to assess performance across different complexity levels [10]. Researchers then applied samples to the different detection platforms under identical buffer and incubation conditions to enable direct comparison. After appropriate incubation and washing steps, they quantified bound ubiquitinated proteins using high-sensitivity detection methods, with ThUBD-HRP employed for the ThUBD platform to ensure consistent detection [10]. The resulting data was analyzed to calculate key performance metrics including sensitivity, dynamic range, and linkage bias for each method [10].
This rigorous comparative approach provided the quantitative data presented in the performance tables, clearly demonstrating the advantages of the ThUBD platform over existing technologies. The workflow also confirmed that the ThUBD-coated plates enabled high-throughput analysis of both global ubiquitination profiles and target-specific ubiquitination status, making the technology suitable for diverse research applications including drug development projects focused on proteolysis-targeting chimeras (PROTACs) [10].
Diagram 1: Experimental workflow comparing ubiquitination detection methods, highlighting key limitations and advantages.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Ubiquitination Studies
| Reagent/Category | Specific Examples | Function and Application |
|---|---|---|
| Ubiquitin Antibodies | P4D1, FK1, FK2 | General ubiquitin detection; immunoblotting and immunofluorescence |
| Linkage-Specific Antibodies | K48-, K63-, M1-specific antibodies | Detection of specific ubiquitin chain linkages; functional studies |
| TUBE Reagents | Commercial TUBE products | Ubiquitinated protein enrichment with moderate affinity and reduced linkage bias vs. antibodies |
| ThUBD Technology | ThUBD-coated plates, ThUBD fusion protein | High-affinity, unbiased ubiquitinated protein capture for sensitive detection |
| Ubiquitinated Protein Standards | Ub-GFP, Ub2-GFP, Ub4-GFP | Method validation and quantification standards |
| Detection Reagents | ThUBD-HRP, anti-His-HRP | Signal generation for quantifying captured ubiquitinated proteins |
| Solid Supports | Corning 3603 96-well plates | High-capacity plates for ThUBD coating and high-throughput applications |
This curated toolkit represents essential reagents for implementing and validating the ubiquitination detection methods discussed in this guide. The ThUBD-coated plates serve as the cornerstone for high-sensitivity, unbiased ubiquitination profiling, particularly valuable for comprehensive studies requiring detection of diverse ubiquitin chain architectures [10]. Ubiquitinated protein standards such as the Ub-GFP series are critical for method validation and establishing quantitative performance metrics [10]. The inclusion of both traditional and advanced reagents enables researchers to select appropriate tools based on their specific experimental requirements, whether for targeted studies of specific ubiquitin linkages or global ubiquitination profiling.
The comprehensive comparison presented in this guide clearly demonstrates that ThUBD-based technology represents a significant advancement in ubiquitination detection methodology, effectively addressing the persistent challenges of low affinity and linkage bias that have limited both antibody-based and conventional UBD-based approaches. The quantitative performance data reveals substantial improvements in key metrics, most notably the 16-fold enhancement in detection sensitivity and 16-fold wider dynamic range achieved by the ThUBD platform compared to TUBE-based methods [10]. These technical advancements directly translate to practical research benefits, enabling detection of low-abundance ubiquitination events and more accurate quantification across diverse sample types.
For researchers and drug development professionals, these methodological advancements open new possibilities for ubiquitination research. The unbiased recognition of ubiquitin chain types enabled by ThUBD technology permits more comprehensive profiling of ubiquitin signaling complexity, potentially revealing novel biological insights that were previously obscured by methodological limitations [10]. The high-throughput capability of the ThUBD-coated plate platform further enhances its utility for drug discovery applications, particularly in the rapidly expanding field of PROTAC development where monitoring target protein ubiquitination is essential [10]. As the ubiquitination research field continues to evolve, methodological improvements that enhance detection sensitivity, reduce linkage bias, and enable high-throughput applications will play an increasingly important role in advancing both basic biological understanding and therapeutic development.
Within the ubiquitin-proteasome system, ubiquitin-binding domains (UBDs) have emerged as indispensable tools for researching protein ubiquitination, a pivotal post-translational modification regulating virtually all cellular processes in eukaryotic cells [4]. The evaluation of enrichment efficiency between UBD-based and antibody-based methods represents a core challenge in proteomics and drug discovery, particularly as researchers seek to understand the complex roles of ubiquitination in disease pathogenesis and therapeutic development [4] [10]. Traditional antibody-based methods, while widely adopted, face significant limitations including linkage bias, limited affinity, and insufficient sensitivity for detecting low-abundance ubiquitinated species within complex proteomes [4] [10] [39]. This comprehensive analysis objectively compares the performance of emerging UBD technologies against conventional antibody-based approaches, providing experimental data and methodologies to guide researchers in selecting optimal tools for ubiquitination studies within the broader context of enriching efficiency evaluation.
Performance Comparison: UBD-Based vs. Antibody-Based Enrichment Methods
Quantitative Performance Metrics
Table 1: Direct performance comparison between ThUBD and TUBE technologies
| Performance Parameter | ThUBD-Coated Plates | TUBE-Coated Plates | Improvement Factor |
|---|---|---|---|
| Detection Sensitivity | 0.625 μg | 10 μg | 16-fold |
| Dynamic Range | Significantly wider | Limited | 16-fold wider linear range |
| Ubiquitin Chain Bias | Unbiased recognition | Linkage-dependent bias | Eliminates bias |
| Affinity for PolyUb Chains | High affinity | Low affinity | Markedly enhanced |
| Throughput Capacity | High-density 96-well plates | Standard 96-well plates | Enhanced throughput |
Table 2: Comparison between UBD-based and antibody-based enrichment methods
| Enrichment Characteristic | UBD-Based Approaches | Antibody-Based Approaches | Key Implications |
|---|---|---|---|
| Recognition Scope | All ubiquitin chain types | Often linkage-specific | ThUBD enables comprehensive profiling |
| Applicability to Native Systems | Suitable for physiological conditions | Requires genetic manipulation for tagging | Antibodies work with clinical samples [4] |
| Specificity Challenges | Minimal non-specific binding | Non-specific binding with histidine-rich/biotinylated proteins [4] | ThUBD reduces background interference |
| Experimental Artifacts | Mimics endogenous Ub interactions | Tagged Ub may alter structure [4] | ThUBD reflects natural ubiquitination |
| Throughput Potential | High-throughput plate formats | Lower throughput (immunoblotting) [4] | ThUBD enables screening applications |
Technical and Practical Considerations
Beyond the quantitative metrics, several technical considerations influence method selection for ubiquitination studies. Antibody-based methods, particularly those using linkage-specific antibodies, remain valuable when studying specific ubiquitin chain types [4]. However, their utility is constrained by the limited availability and high cost of quality antibodies, with systematic assessments revealing that only 45% (70/157) of tested antibodies successfully enriched their expected target proteins, and among these, 84% co-enriched other proteins due to sequence homology or abundance effects [39].
UBD-based technologies, particularly the Tandem Hybrid Ubiquitin Binding Domain (ThUBD) system, address these limitations through engineered solutions that combine the advantages of different ubiquitin-binding domains [10]. This integration yields both high affinity for polyubiquitinated proteins and unbiased recognition across all ubiquitin chain types, representing a significant advancement over traditional Tandem Ubiquitin Binding Entities (TUBEs) which exhibit inherent linkage bias and limited binding capacity [10].
Experimental Platforms and Methodologies
ThUBD-Coated High-Density 96-Well Plate Platform
Platform Design and Optimization
The ThUBD-coated plate technology represents a sophisticated high-throughput platform for ubiquitination research. The methodology involves coating 1.03 μg ± 0.002 of ThUBD fusion protein on Corning 3603-type 96-well plates, enabling specific binding to approximately 5 pmol of polyubiquitin chains [10]. This carefully optimized system demonstrates a remarkable 16-fold enhancement in detection sensitivity compared to conventional TUBE-coated plates, with the capability to capture ubiquitinated proteins from complex proteome samples even at low input levels of 0.625 μg [10].
The experimental workflow begins with coating high-capacity 96-well plates with ThUBD under optimized conditions, followed by incubation with proteome samples. After thorough washing with specifically formulated buffers, the captured ubiquitinated proteins are detected and quantified, typically using ThUBD-HRP conjugates for sensitive measurement [10]. This streamlined process enables both global ubiquitination profiling and target-specific ubiquitination status assessment, making it particularly valuable for dynamic monitoring of ubiquitination in Proteolysis-Targeting Chimeras (PROTACs) drug development [10].
Experimental Validation
Rigorous validation experiments demonstrate that ThUBD-coated plates can efficiently capture polyubiquitinated proteins ranging from ubiquitin-green fluorescent protein (Ub-GFP) fusion constructs to endogenous ubiquitinated proteins from HEK293T cell lysates [10]. The system shows unprecedentedly low non-specific binding while maintaining robust signal-to-noise ratios across diverse sample types, establishing it as a reliable platform for both qualitative detection and precise quantification of ubiquitination signals.
Mass Spectrometry-Based Ubiquitinome Analysis
Ubiquitin Tagging-Based Enrichment
A primary methodology for ubiquitinome analysis involves tagging-based enrichment approaches, where Ub containing affinity tags (such as His, Flag, HA, or Strep tags) is expressed in living cells, enabling purification of ubiquitinated substrates using commercially available resins including Ni-NTA for His tag and Strep-Tactin for Strep-tag [4]. Following purification and tryptic digestion of ubiquitinated proteins, ubiquitination sites are determined through mass spectrometry analysis by identifying the characteristic 114.04 Da mass shift on modified lysine residues [4].
This approach was pioneered in 2003 by Peng et al., who identified 110 ubiquitination sites on 72 proteins from Saccharomyces cerevisiae [4]. Subsequent refinements, including the stable tagged Ub exchange (StUbEx) cellular system developed by Akimov et al., significantly enhanced identification efficiency, yielding 277 unique ubiquitination sites on 189 proteins in HeLa cells [4]. While this method provides a relatively low-cost approach for screening ubiquitinated substrates, limitations include co-purification of histidine-rich or endogenously biotinylated proteins, potential structural alterations from tagged Ub, and limited application in animal or patient tissues where genetic manipulation is infeasible [4].
Immunoprecipitation-Mass Spectrometry (IP-MS) Workflow
For antibody-based ubiquitin enrichment, researchers employ a well-established IP-MS workflow that begins with antibody covalent coupling to magnetic polystyrene beads, followed by incubation with plasma or cell lysate samples [39]. After target enrichment and washing, proteins are digested on beads and resulting peptides are analyzed by liquid chromatography-mass spectrometry (LC-MS) [39]. Data processing typically involves MaxLFQ for label-free quantification, with subsequent z-score analysis to rank proteins specifically enriched by each antibody compared to population-level background [39].
Systematic assessments using this workflow have revealed significant specificity challenges, with only 45% of tested antibodies effectively enriching their expected target proteins, and among these, 84% co-enriching other proteins primarily due to sequence homology or abundance effects [39]. This highlights the critical importance of thorough antibody validation when employing immunological enrichment methods.
Diagram 1: Comparative workflows for UBD-based and antibody-based ubiquitin enrichment methods. The ThUBD-coated plate platform (red) offers a streamlined approach with minimal non-specific binding, while antibody-based IP (green) involves more complex processing with potential specificity challenges.
Research Reagent Solutions Toolkit
Table 3: Essential research reagents for ubiquitination studies
| Reagent / Tool | Primary Function | Specific Application Examples |
|---|---|---|
| ThUBD Fusion Protein | High-affinity, unbiased ubiquitin chain capture | Coating high-density plates; ubiquitinated protein enrichment from complex samples [10] |
| Linkage-Specific Ub Antibodies | Selective enrichment of specific ubiquitin chain types | Studying K48-linked degradation signaling; K63-linked NF-κB pathway analysis [4] |
| TUBE (Tandem Ubiquitin Binding Entity) | General polyubiquitin enrichment with some linkage bias | Traditional ubiquitination pull-downs; proteasome interaction studies [10] |
| Tagged Ubiquitin Constructs | Expression-based ubiquitination profiling | Identification of ubiquitination sites; monitoring ubiquitination dynamics in live cells [4] |
| Affinity Resins (Ni-NTA, Strep-Tactin) | Purification of tagged ubiquitinated proteins | Isolation of His-tagged ubiquitin conjugates; Strep-tagged ubiquitin proteomics [4] |
| DUB Inhibitors | Preservation of ubiquitination signals | Preventing deubiquitination during sample preparation; stabilizing labile ubiquitin conjugates |
| Ubiquitin Activating Enzyme Inhibitors | Disruption of ubiquitination cascades | Control experiments; validating specificity of ubiquitination detection methods |
Technical Guidelines for Method Selection
Context-Dependent Recommendation Framework
The optimal method selection for ubiquitination studies depends heavily on specific research objectives and experimental constraints. For comprehensive ubiquitinome profiling requiring unbiased detection across all chain types, the ThUBD-platform demonstrates superior performance, particularly when working with limited sample material where its enhanced sensitivity provides significant advantages [10]. When investigating specific ubiquitin linkage types with established, well-validated antibodies, traditional immunoaffinity approaches may yield satisfactory results, though researchers should implement rigorous controls to verify specificity [39].
For discovery-phase research aiming to identify novel ubiquitination events without prior knowledge of targeted biomolecules, the integrated computational and experimental iLaMA (integrated Law of Mass Action) approach offers powerful capabilities, having successfully identified approximately 10^5 antibody sequences encoding specificity for tumor cell surface receptors expressed across a remarkable range of 10^3–10^6 receptors per cell [40].
Emerging Applications and Future Directions
The evolving landscape of ubiquitination research continues to generate new applications for optimized UBD technologies. In targeted protein degradation drug discovery, particularly for PROTAC development, the ThUBD-coated plate platform enables efficient monitoring of ubiquitination status, providing critical technical support for this promising therapeutic strategy [10]. Additionally, multi-omics approaches integrating ubiquitinome data with genomic and transcriptomic profiles are advancing our understanding of molecules like Ubiquitin D (UBD/FAT10), with pan-cancer analyses revealing its overexpression in 29 cancer types and correlation with poor prognosis, highlighting the clinical relevance of advanced ubiquitination detection methods [41].
Future methodology developments will likely focus on enhancing sensitivity for single-cell ubiquitination analysis, improving spatial resolution of ubiquitination events within cellular compartments, and developing more sophisticated computational integration of ubiquitinome data with other signaling network information. The continued optimization of UBD performance through protein engineering approaches promises to further expand the capture capacity and specificity achievable in ubiquitination research.
Protein ubiquitination is a crucial post-translational modification regulating diverse cellular functions, including protein degradation, DNA repair, and immune signaling [42] [4]. However, proteome-wide profiling of ubiquitinated proteins presents significant technical challenges due to their low abundance, the diversity of ubiquitin chain linkages, and the transient nature of these modifications [42] [4]. Researchers primarily employ two strategic approaches for ubiquitin enrichment: antibody-based methods and ubiquitin-binding domain (UBD)-based technologies. Each approach faces distinct challenges in high-throughput settings where scalability and reproducibility are paramount for generating robust, biologically relevant data.
Antibody-based methods have long been the workhorse of ubiquitin research, but concerns about characterization and reproducibility have prompted the exploration of alternative UBD-based methods [43]. Simultaneously, UBD-based tools have evolved from single low-affinity domains to sophisticated multi-domain constructs with enhanced capabilities [42] [11]. This comparison guide objectively evaluates the performance of these competing methodologies within high-throughput research environments, providing experimental data and protocols to inform researchers' selection of appropriate ubiquitin enrichment strategies.
Methodological Foundations: Core Technologies for Ubiquitin Enrichment
Antibody-Based Enrichment Methods
Antibody-based approaches utilize immunoprecipitation with antibodies that recognize ubiquitin or ubiquitin remnants. The primary categories include:
- Pan-ubiquitin antibodies (e.g., FK1, FK2, P4D1) that recognize all ubiquitin linkages and are used for enriching ubiquitinated proteins [4].
- Linkage-specific antibodies that target particular ubiquitin chain topologies (e.g., K48-, K63-, M1-linked chains) [4].
- DiGly remnant antibodies that enrich tryptic peptides containing the GlyGly modification left on ubiquitinated lysines after proteolytic digestion [42].
A significant challenge in antibody-based research is what has been termed the "antibody characterization crisis," with an estimated 50% of commercial antibodies failing to meet basic characterization standards [43]. This problem contributes to substantial financial losses estimated at $0.4-1.8 billion annually in the United States alone and compromises experimental reproducibility [43].
UBD-Based Enrichment Technologies
UBD-based methods utilize recombinant ubiquitin-binding domains to capture ubiquitinated proteins:
- Single UBDs: Early approaches used individual ubiquitin-binding domains but suffered from low affinity (Kd values in micromolar range) [42].
- TUBEs (Tandem Ubiquitin-Binding Entities): Multiple UBDs fused together to increase avidity for polyubiquitin chains [42] [4].
- Engineered High-Affinity UBDs: Novel tools like OtUBD (derived from Orientia tsutsugamushi deubiquitylase) with exceptional binding affinity (Kd ≈ 5 nM) [42].
- Hybrid UBDs: Artificially designed constructs like ThUBDs (Tandem Hybrid UBDs) combining different UBD types for enhanced performance [11].
Table 1: Key Ubiquitin-Binding Domains and Their Properties
| UBD Type | Source | Affinity | Key Features | Reference |
|---|---|---|---|---|
| OtUBD | Orientia tsutsugamushi | Kd ≈ 5 nM | Binds I44 hydrophobic patch; detects mono- and polyubiquitination | [42] |
| Ubiquilin-1 UBA | Human | Micromolar range | Basis for 4xUBA TUBEs; prefers polyubiquitin | [44] |
| ThUBDs (ThUDQ2, ThUDA20) | Engineered | Markedly higher than natural UBDs | Unbiased high affinity to all lysine-linked chains | [11] |
| DSK2p-derived UBA | Engineered | High affinity | Component of hybrid ThUBDs | [11] |
Experimental Comparison: Performance Metrics in High-Throughput Applications
Efficiency in Capturing Diverse Ubiquitination Types
A critical differentiator between enrichment methods is their ability to detect various ubiquitination forms:
Monoubiquitination Detection: OtUBD demonstrates exceptional capability in preserving and enriching monoubiquitylated species, as evidenced by experiments with histone H2B in yeast, where it preserved monoubiquitylated H2B comparably to the DUB inhibitor NEM [42]. In contrast, TUBEs showed limited efficiency for monoubiquitylated proteins, with complete loss of H2B-ubiquitin signal in experimental conditions [42]. This is significant given that over 50% of ubiquitylated proteins are exclusively monoubiquitylated in some mammalian cell types [42].
Polyubiquitin Chain Linkage Recognition: Comprehensive evaluation of ThUBDs revealed nearly unbiased high affinity to all seven lysine-linked ubiquitin chains, making them superior for global ubiquitome profiling [11]. Similarly, OtUBD efficiently purified both monoubiquitylated and polyubiquitylated substrates from yeast and human samples [42]. Linkage-specific antibodies remain valuable for studying particular chain types but require multiple parallel experiments for comprehensive analysis.
Non-Canonical Ubiquitination Detection: Unlike diGly antibody approaches that are restricted to lysine modifications, UBD-based methods can potentially detect non-canonical ubiquitination on serine, threonine, cysteine, or N-terminal residues [42]. This represents a significant advantage for discovering novel ubiquitination types.
Scalability and Throughput Considerations
High-throughput applications demand methods that are scalable, cost-effective, and compatible with automation:
Reagent Production and Cost: UBD-based tools like OtUBD and ThUBDs are produced recombinantly in bacterial systems, offering low-cost, economical alternatives to antibodies [42] [11]. The production process is standardized and scalable, avoiding batch-to-batch variability common with antibody production [43].
Compatibility with Automated Systems: Recombinant UBD constructs with tags such as GST or MBP seamlessly integrate into high-throughput purification workflows using automated liquid handlers and affinity resin processing [42] [11]. Antibody-based methods require careful optimization to maintain antibody integrity in automated systems.
Processing Time: UBD-based affinity purifications typically require 2-4 hours incubation followed by wash and elution steps, comparable to antibody-based immunoprecipitations [42]. However, the elimination of need for epitope-tagged ubiquitin overexpression in UBD approaches saves days of cell culture time.
Table 2: Throughput and Scalability Comparison
| Parameter | Antibody-Based Methods | UBD-Based Methods |
|---|---|---|
| Reagent Production | Hybridoma/ recombinant expression | Bacterial recombinant expression |
| Batch Consistency | Variable (50% fail characterization) [43] | High (defined sequence) |
| Cost per Enrichment | High (antibody production) | Low (recombinant protein) |
| Automation Compatibility | Moderate (sensitivity to handling) | High (robust constructs) |
| Multiplexing Potential | Limited | Higher (multiple tags possible) |
| Process Standardization | Challenging | Straightforward |
Reproducibility and Experimental Variance
Reproducibility concerns significantly impact antibody-based methods. A comprehensive analysis of commercial antibodies revealed that inadequate characterization leads to unreliable results, with consequences including misleading conclusions in clinical patient trials [43]. UBD-based tools with defined amino acid sequences and standardized production minimize lot-to-lot variability.
Experimental data demonstrates that OtUBD provides consistent enrichment across biological replicates in both yeast and human tissue culture samples [42]. Similarly, ThUBDs showed reproducible identification of ubiquitination sites, with 362 sites identified in yeast and 1,125 in mammalian cells [11].
Experimental Protocols for High-Throughput Applications
OtUBD Affinity Purification Protocol
Materials:
- Recombinant MBP-OtUBD or MBP-3xOtUBD fusion protein
- Amylose resin
- Lysis buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, protease inhibitors
- Wash buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% NP-40
- Elution buffer: Wash buffer + 10-20 mM maltose
Methodology:
- Couple purified MBP-OtUBD fusion protein to amylose resin (2-4 hours, 4°C)
- Prepare cell lysates in lysis buffer (supplement with NEM or other DUB inhibitors)
- Incubate lysate with resin-bound OtUBD (2-3 hours, 4°C)
- Wash resin extensively with wash buffer (5-10 column volumes)
- Elute bound ubiquitinated proteins with elution buffer
- Process eluates for Western blot or mass spectrometry analysis
Validation: Researchers validated this protocol by profiling the ubiquitylome and ubiquitin-associated proteome of Saccharomyces cerevisiae, identifying potential substrates for E3 ligases Bre1 and Pib1 [42]. The method efficiently enriched both monoubiquitylated and polyubiquitylated species.
ThUBD Enrichment Protocol for Mass Spectrometry
Materials:
- GST-ThUBD fusion proteins (ThUDQ2 or ThUDA20)
- Glutathione sepharose
- Lysis buffer: 50 mM HEPES (pH 7.5), 150 mM NaCl, 1% Triton X-100, 0.5 mM DTT, protease inhibitors
- Wash buffer: 50 mM HEPES (pH 7.5), 300 mM NaCl, 0.5% Triton X-100
- Elution buffer: Wash buffer + 20 mM reduced glutathione
Methodology:
- Immobilize GST-ThUBD on glutathione sepharose
- Prepare cell lysates (use DUB inhibitors to preserve ubiquitination)
- Incubate lysate with ThUBD resin (3-4 hours, 4°C)
- Wash with high-salt wash buffer to reduce non-specific binding
- Elute with glutathione-containing buffer
- Denature, reduce, alkylate, and digest enriched proteins with trypsin
- Analyze peptides by LC-MS/MS
Performance Metrics: This protocol identified 1,092 putative ubiquitinated proteins from yeast and 7,487 from mammalian cells, with 362 and 1,125 ubiquitination sites confirmed, respectively [11].
Diagram 1: Experimental Workflow for Ubiquitin Enrichment Methods. This diagram illustrates parallel pathways for antibody-based and UBD-based ubiquitin enrichment, converging on common downstream analytical techniques.
Research Reagent Solutions: Essential Materials for Ubiquitin Enrichment
Table 3: Key Research Reagents for Ubiquitin Enrichment Studies
| Reagent | Type | Function | Applications | Considerations |
|---|---|---|---|---|
| OtUBD | Recombinant UBD | High-affinity ubiquitin binding | Enrichment of mono- and polyubiquitinated proteins | Bacterial expression; MBP or GST tags [42] |
| ThUBDs (ThUDQ2, ThUDA20) | Engineered tandem UBDs | Enhanced affinity for diverse ubiquitin chains | Global ubiquitome profiling by MS | Unbiased recognition of linkage types [11] |
| 4xUBA TUBEs | Tandem UBDs | Polyubiquitin chain enrichment | Proteasomal substrate identification | Lower affinity for monoubiquitination [42] [44] |
| FK2/P4D1 Antibodies | Pan-ubiquitin antibodies | Recognize ubiquitinated proteins | Immunoprecipitation, Western blot | Characterization variability concerns [4] [43] |
| Linkage-Specific Antibodies | Specific ubiquitin chain antibodies | Target particular ubiquitin linkages | Studying specific ubiquitin signals | Limited to predetermined linkages [4] |
| diGly Remnant Antibodies | Modification-specific antibodies | Enrich ubiquitinated peptides | Ubiquitination site mapping by MS | Limited to lysine ubiquitination [42] |
The comparative analysis of UBD-based versus antibody-based methods for ubiquitin enrichment reveals a complex landscape where method selection must be guided by specific research goals and throughput requirements.
For high-throughput applications requiring comprehensive ubiquitome profiling, UBD-based methods—particularly novel tools like OtUBD and engineered ThUBDs—offer significant advantages in scalability, reproducibility, and cost-effectiveness. Their ability to detect diverse ubiquitination types, including monoubiquitination and non-canonical modifications, provides a more complete picture of the ubiquitin landscape.
Antibody-based methods remain valuable for targeted studies of specific ubiquitin chain linkages or when working with limited sample amounts, though researchers must prioritize thorough antibody characterization and include appropriate controls to ensure reproducibility.
Future directions in ubiquitin enrichment technology will likely focus on further engineering of UBDs with enhanced specificity and affinity, development of automated high-throughput platforms, and integration with emerging proteomics technologies. As the field addresses current challenges in scalability and reproducibility, these advanced reagents will continue to drive discoveries in ubiquitin biology and therapeutic development.
Diagram 2: Method Selection Guide for Ubiquitin Enrichment. This decision-support diagram outlines scenarios where antibody-based or UBD-based methods are most appropriate, helping researchers select optimal approaches for their specific needs.
Critical Buffer and Coating Conditions for Maximizing Target Protein Recovery
The efficacy of proteomic research and biopharmaceutical development hinges on the successful recovery of target proteins from complex biological samples. This process is critically dependent on the initial steps of protein extraction, enrichment, and immobilization, which are governed by the choice of buffers and solid-phase coatings. Within the broader context of evaluating the enrichment efficiency of ubiquitin-binding domain (UBD)-based versus antibody-based methods, optimizing these initial conditions is not merely a procedural detail but a fundamental determinant of experimental success. Inefficient protein recovery can lead to substantial quantitative biases, masking true biological signals and compromising the reliability of downstream analyses [45]. This guide provides a objective comparison of critical methodologies and reagents, focusing on their performance in maximizing the recovery of target proteins, with a specific emphasis on applications within ubiquitination research. The data and protocols presented herein are designed to empower researchers to make informed decisions that enhance yield, purity, and functional integrity across various sample types, including the challenging yet invaluable formalin-fixed paraffin-embedded (FFPE) tissues.
Methodological Comparison: UBD-Based vs. Antibody-Based Enrichment
The selection of an enrichment strategy is a primary decision point in targeted proteomics. Two predominant methods are employed for isolating specific proteins or post-translationally modified species, such as ubiquitinated targets: antibody-based enrichment and UBD-based affinity capture. The following section provides a detailed, data-driven comparison of their performance characteristics, advantages, and limitations.
Table 1: Performance Comparison of Ubiquitinated Protein Enrichment Methods
| Feature | Antibody-Based Methods | UBD-Based Methods |
|---|---|---|
| Basis of Recognition | Immunoreactivity with specific epitopes or ubiquitin chain linkages [4]. | Reversible protein-protein interaction with ubiquitin moieties [4] [11]. |
| Typical Reagents | Linkage-specific anti-ubiquitin antibodies (e.g., K48-, K63-specific) [4]. | Engineered tandem hybrid UBDs (ThUBDs), single UBA domains, tandem-repeated UBDs [4] [11]. |
| Key Advantage | High specificity for defined ubiquitin linkages; applicable to tissue samples without genetic manipulation [4]. | Broader affinity for various ubiquitin chain linkages; avoids need for tagged ubiquitin overexpression [11]. |
| Key Limitation | Potential for high cost, non-specific binding, and loss of immunoreactivity in stored FFPE samples [45] [4]. | Lower affinity of single UBDs; requires careful engineering for high-affinity binding [4] [11]. |
| Typical Application | Immunoblotting, immunohistochemistry (IHC), enrichment of proteins with specific ubiquitin linkages from tissues [4]. | Proteome-wide profiling of ubiquitinated proteins in cells; identification of ubiquitination sites via MS [4] [11]. |
| Reported Performance | Enabled identification of 96 ubiquitination sites from MCF-7 cells [4]. | ThUBDs identified 1092 (yeast) to 7487 (mammalian) ubiquitinated proteins [11]. |
The experimental data indicates that UBD-based methods, particularly engineered ThUBDs, can achieve markedly higher affinity and broader specificity for diverse ubiquitin chains compared to naturally occurring UBDs [11]. This makes them a powerful tool for unbiased ubiquitin proteomics. Conversely, antibody-based methods are susceptible to signal loss in long-term stored FFPE tissues, a phenomenon attributed to loss of immunoreactivity rather than protein degradation [45]. This is a critical consideration for retrospective clinical studies. Furthermore, while antibody-based methods can be exquisitely specific, their utility in discovery-based proteomics may be limited by availability, cost, and potential for non-specific binding [4].
Critical Buffer Formulations for Protein Extraction and Recovery
The buffer system used for protein extraction is a pivotal factor for yield and quality. The optimal formulation must efficiently solubilize the target protein, reverse cross-links (in the case of FFPE samples), and maintain protein stability, all while being compatible with downstream enrichment and analysis steps like mass spectrometry (MS).
Table 2: Comparison of Protein Extraction Buffer Efficacy from FFPE Tissues
| Extraction Buffer | Key Components | Reported Performance / Advantage | Key Limitation |
|---|---|---|---|
| Zwittergent-Based Buffer | 0.2% Zwittergent 3-16, 10 mM Tris, 1 mM EDTA [46]. | Most efficient for identifying peptides and proteins across multiple rat tissues; well-compatible with MS [46]. | Limited lytic strength for certain insoluble proteins, like membrane proteins [46]. |
| Surfactant Cocktail (f-SEPOD) | Proprietary surfactant cocktail [45]. | Provides high/reproducible recovery of target signature peptides from FFPE tissues; enables accurate LC-MS quantification [45]. | Requires optimization for different tissue types [45]. |
| SDS-Based Buffer | 4% SDS, 100 mM Tris, 100 mM DTT, pH 8.0 [46]. | Effective for protein unfolding and retrieval; identified ~2000 proteins from renal cell carcinoma FFPE samples [46]. | Incompatible with MS; requires detergent removal prior to analysis [46]. |
| SDS-Based Buffer with PEG20000 | 4% SDS, 100 mM Tris, 100 mM DTT, 0.5% PEG20000, pH 8.0 [46]. | PEG20000 acts as a carrier, improving identification of peptides and proteins from low-protein samples [46]. | Benefit is limited to tissues with submicrogram to microgram protein levels [46]. |
| Urea-Based Buffer | 8 M Urea, 2 M Thiourea, 4% CHAPS, 65 mM DTT [46]. | MS-compatible; no need for detergent removal [46]. | Limited lytic strength; leaves certain proteins insoluble [46]. |
The comparative study of FFPE tissue extraction demonstrates that Zwittergent 3-16-based buffer consistently outperformed other formulations, including SDS and urea-based systems, in the number of peptides and proteins identified across various tissue types [46]. For targeted, absolute quantification, the surfactant cocktail-based f-SEPOD procedure was developed specifically to address the challenge of quantitative signature peptide recovery from FFPE tissues, a prerequisite for accurate LC-MS analysis [45]. It is also critical to note that the choice of calibration standard is paramount; using FFPE-treated calibration standards instead of conventionally spiked standards was shown to prevent substantial negative quantitative biases [45].
Detailed Experimental Protocol: f-SEPOD for FFPE Tissues
The following protocol, adapted from the highly accurate LC-MS-based quantification pipeline, details the steps for maximizing protein recovery from FFPE tissues [45].
- Deparaffinization and Rehydration: Cut sections from the FFPE block and incubate through three changes of xylene (5 min each). Rehydrate through a series of graded ethanol (100%, 90%, 80%, 70% for 1 min each) and incubate in distilled water for at least 30 min.
- Tissue Homogenization: Scrape tissue sections from the slides and pool them. For optimal homogenization, pulverize the pooled FFPE tissue pieces into a fine powder under liquid nitrogen.
- Surfactant Cocktail-Based Extraction and Digestion: Homogenously mix the tissue powder with the proprietary surfactant cocktail-based extraction buffer. The specific formulation of the "f-SEPOD" buffer is optimized to reverse formalin-induced cross-links efficiently.
- Heat-Assisted Extraction: Heat the homogenate at 100°C for 20 minutes, followed by incubation at 60°C for 2 hours. This heat step is critical for reversing cross-links.
- Sonication: Subject the sample to 40 minutes of sonication in a water bath to further aid in protein solubilization.
- Protein Reduction and Alkylation: Reduce the protein extracts in 20 mM DTT at 37°C for 60 minutes. Subsequently, alkylate the samples with 25 mM iodoacetamide in the dark for 45 minutes at room temperature.
- Clean-up and Digestion: Centrifuge the extracts (21,000×g for 10 min at 4°C) to pellet insoluble debris. Transfer the supernatant to a spin ultrafiltration unit (10 kDa MWCO) for detergent removal and buffer exchange into 25 mM ammonium bicarbonate (NH₄HCO₃). Perform tryptic digestion on the retained proteins.
- LC-MS Analysis: The resulting peptides, including the target signature peptides, are now ready for highly accurate and precise absolute quantification by Liquid Chromatography-Multiple Reaction Monitoring-Mass Spectrometry (LC-MRM-MS) [45].
The Scientist's Toolkit: Essential Research Reagents
The following table catalogues key reagents and materials cited in the featured research, providing an overview of their primary functions in protein recovery workflows.
Table 3: Key Research Reagent Solutions for Protein Recovery
| Reagent / Material | Function in Protein Recovery | Reference |
|---|---|---|
| Zwittergent 3-16 | Detergent for efficient protein extraction from FFPE tissues; MS-compatible. | [46] |
| Surfactant Cocktail (f-SEPOD) | Optimized mixture for high-yield recovery of signature peptides from FFPE tissues for LC-MS. | [45] |
| PEG 20000 | High molecular weight carrier that enhances protein identification in low-yield samples. | [46] |
| Tandem Hybrid UBDs (ThUBDs) | Engineered affinity ligands for high-affinity, broad-specificity enrichment of ubiquitinated proteins. | [11] |
| Capto MMC | Multimodal chromatographic resin combining ion-exchange and hydrophobic interactions for protein purification. | [47] [48] |
| Linkage-specific Ub Antibodies | Immunoaffinity reagents for enriching proteins with specific ubiquitin chain linkages (e.g., K48, K63). | [4] |
| Coating Buffers | Optimize adsorption and stabilize tertiary structure of antibodies/antigens on solid surfaces (e.g., ELISA plates). | [49] |
Workflow and Relationship Diagrams
The following diagram illustrates the logical sequence and decision points for selecting the optimal buffer and enrichment strategy based on sample type and research goals.
Diagram 1: Decision Workflow for Protein Recovery and Enrichment. This chart outlines the critical decision points for selecting an optimal buffer and enrichment method based on sample type and research objectives, guiding researchers from sample collection to downstream analysis.
The interactions between proteins, buffers, and purification resins are complex. The following diagram summarizes the key factors that govern protein adsorption and recovery on a multimodal chromatographic resin, which exemplifies the multi-faceted nature of downstream processing.
Diagram 2: Key Factors Governing Multimodal Chromatography. This diagram visualizes the complex interplay between critical process parameters (buffer conditions), critical material attributes (protein properties), and the resulting binding mechanisms that determine the success of a purification run.
Navigating Artifacts and Non-Specific Binding in Complex Samples
Protein ubiquitination, a crucial post-translational modification, regulates diverse cellular functions including protein degradation, DNA repair, and immune responses [4]. The detection and accurate quantification of ubiquitination signals are fundamental to advancing our understanding of cellular physiology and pathology, particularly in drug development areas such as Proteolysis-Targeting Chimeras (PROTACs) [10]. However, researchers face significant methodological challenges in this endeavor, primarily concerning non-specific binding and the generation of analytical artifacts that can compromise data integrity. These challenges are particularly pronounced when working with complex biological samples where target proteins exist within a milieu of abundant non-target molecules [50] [4].
The scientific community has primarily relied on two methodological approaches for ubiquitin detection: antibody-based methods and ubiquitin-binding domain (UBD)-based technologies. Antibody-based methods utilize immunoglobulins raised against ubiquitin or specific ubiquitin chain linkages, while UBD-based approaches exploit natural protein domains that recognize and bind ubiquitin motifs [4] [10]. Each strategy presents distinct advantages and limitations concerning specificity, affinity, linkage bias, and susceptibility to experimental artifacts. This guide provides an objective comparison of these methodologies, focusing on their respective capabilities to minimize analytical confounders while providing reliable detection and quantification of ubiquitination events in complex sample types.
Methodological Principles and Technical Challenges
Antibody-Based Enrichment Methods
Antibody-based methods for ubiquitin detection include techniques such as immunoaffinity chromatography, immunoprecipitation (IP), and co-immunoprecipitation (Co-IP) [38]. These methods rely on the specific binding affinity between an antibody and its target epitope on ubiquitin or ubiquitin chains. The fundamental principle involves immobilizing antibodies on a solid support matrix, such as protein A/G agarose or magnetic beads, which then capture ubiquitinated proteins from complex samples [38]. After washing away non-specifically bound proteins, the target ubiquitinated proteins are eluted for downstream analysis.
A significant limitation of conventional antibody-based methods is their inherent linkage bias. Available antibodies often exhibit preferential affinity for specific ubiquitin chain linkages (e.g., K48 or K63), while demonstrating reduced recognition of atypical chains (K6, K11, K27, K29, K33) or linear ubiquitin [4] [10]. This bias can create a distorted representation of the cellular ubiquitin landscape. Additionally, antibody-based methods face challenges related to low-affinity antibodies targeting the highly conserved ubiquitin protein itself, potentially leading to inefficient capture and reduced detection sensitivity [10]. Non-specific binding remains a persistent concern, where antibodies may interact with non-target proteins through electrostatic, hydrophobic, or Fc-receptor interactions, particularly in complex samples like cell lysates or plasma [50] [51]. For instance, in flow cytometry, fluorochrome-conjugated antibodies can exhibit non-specific binding to Fc-receptors, creating artifacts that require blocking strategies with antibodies like 2.4G2 to mitigate [51].
Ubiquitin-Binding Domain (UBD)-Based Methods
UBD-based methodologies exploit natural protein domains that have evolved to recognize and bind ubiquitin motifs. Early implementations utilized single UBDs but suffered from low affinity, limiting their utility for efficient enrichment [4]. Technological advancements have led to the development of engineered tandem hybrid ubiquitin-binding domains (ThUBDs) that combine multiple UBDs to achieve unbiased recognition and high affinity for diverse ubiquitin chain types [10].
The principle behind UBD-based enrichment involves expressing and purifying these engineered domains as fusion proteins, which are then immobilized on solid supports such as agarose beads or the surfaces of multi-well plates [10]. These platforms capture ubiquitinated proteins from complex mixtures through high-affinity interactions with ubiquitin chains. Compared to antibody-based methods, well-designed UBD approaches demonstrate superior capability to recognize all ubiquitin linkage types without preference, providing a more comprehensive view of the ubiquitinome [10]. The enhanced binding affinity of engineered ThUBDs allows for more efficient capture of ubiquitinated proteins, even from dilute solutions or samples with low target abundance.
Both methodological approaches must contend with several sources of artifacts and non-specific interactions:
- Matrix effects: The solid support matrix (dextran, agarose, plastic surfaces) can interact non-specifically with sample components through hydrophobic or charged interactions [50].
- Cellular interferents: In flow cytometry, intracellular biotin can bind streptavidin-conjugated reagents, while highly charged fluorochromes like FITC can exhibit electrostatic binding to cellular components [51].
- Sample preparation artifacts: In mass spectrometry analyses, adduct formation with metal ions (sodium, potassium, calcium) can create artifact peaks that complicate interpretation [52].
- Protein degradation: Post-mortem enzymatic activity can rapidly degrade ubiquitination signals if samples are not properly preserved [53].
Strategies to minimize these artifacts include using reference surfaces [50], modifying buffer conditions (increased salt, detergents) [50], implementing blocking steps [51], and ensuring rapid sample processing [53].
Comparative Performance Analysis
Direct Method Comparison: ThUBD vs. Antibody-Based Platforms
Recent technological advancements have led to the development of high-throughput detection platforms using both UBD and antibody-based principles. The following table summarizes experimental data comparing the performance of ThUBD-coated plates with traditional TUBE (Tandem Ubiquitin Binding Entity)-coated plates, which represent two generations of UBD-based technology, along with general antibody-based approaches.
Table 1: Performance comparison of ubiquitin detection methods
| Parameter | ThUBD-Coated Plates | TUBE-Coated Plates | Traditional Antibody-Based Methods |
|---|---|---|---|
| Detection Sensitivity | 0.625 μg | 10 μg (16-fold lower than ThUBD) | Variable; often limited by antibody affinity |
| Linear Dynamic Range | 16-fold wider than TUBE | Limited | Restricted |
| Linkage Bias | No bias toward any ubiquitin chain type | Bias toward specific linkages | Strong bias toward specific linkages |
| Capture Specificity | High specificity for polyubiquitin chains | Moderate | Variable; dependent on antibody quality |
| Throughput Capability | High (96-well format) | Moderate | Low to moderate |
| Non-Specific Binding | Low with optimized washing | Moderate | Can be significant without proper controls |
| Major Limitations | Requires specialized ThUBD production | Limited affinity, linkage bias | Linkage bias, high-quality antibody dependency |
The experimental data reveals that ThUBD-coated plates demonstrate significantly enhanced sensitivity, detecting ubiquitinated proteins at quantities as low as 0.625 μg compared to 10 μg for TUBE-based methods—representing a 16-fold improvement [10]. This enhanced sensitivity is particularly valuable when working with limited clinical samples or low-abundance ubiquitinated targets. Furthermore, ThUBD technology exhibits a 16-fold wider linear range for capturing polyubiquitinated proteins from complex proteome samples, enabling more accurate quantification across a broader concentration spectrum [10].
A critical distinction lies in linkage recognition capability. ThUBD-based platforms show no bias toward any ubiquitin chain type, enabling unbiased enrichment and detection of all ubiquitin linkages including K48, K63, and atypical chains [10]. In contrast, both TUBE and antibody-based methods exhibit significant linkage preferences that can skew experimental results and provide an incomplete picture of ubiquitination status [4] [10].
Experimental Protocols for Critical Comparisons
Protocol: ThUBD Plate Assay for Ubiquitination Detection
- Plate Coating: Coat Corning 3603-type 96-well plates with 1.03 μg ± 0.002 of ThUBD protein in carbonate-bicarbonate buffer (pH 9.6) and incubate overnight at 4°C [10].
- Blocking: Block plates with 5% bovine serum albumin (BSA) in PBS for 2 hours at room temperature to minimize non-specific binding.
- Sample Application: Apply complex proteome samples (cell lysates, tissue homogenates) diluted in binding buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% NP-40, 1 mM DTT) to wells.
- Incubation and Washing: Incubate for 2 hours at room temperature with gentle agitation. Wash three times with optimized washing buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.05% Tween-20) to remove non-specifically bound proteins.
- Detection: Incubate with ThUBD-HRP conjugate for 1 hour. After final washing, develop with chemiluminescent substrate and measure signal intensity [10].
Protocol: Immunoprecipitation for Ubiquitination Analysis
- Antibody Immobilization: Incubate 1-5 μg of anti-ubiquitin antibody (e.g., P4D1, FK1, FK2, or linkage-specific antibodies) with protein A/G agarose beads for 2 hours at 4°C [4] [38].
- Blocking: Block beads with 2-5% BSA to reduce non-specific binding.
- Sample Preparation: Prepare cell lysates in IP lysis buffer (40 mM HEPES, pH 7.4, 120 mM NaCl, 1 mM EDTA, 0.3% CHAPS, 10 mM pyrophosphate, 10 mM glycerophosphate, 50 mM NaF) supplemented with protease inhibitors and 10 mM N-ethylmaleimide to inhibit deubiquitinases.
- Immunoprecipitation: Incubate pre-cleared lysates with antibody-bound beads overnight at 4°C with rotation.
- Washing: Wash beads three times with lysis buffer containing 300-500 mM NaCl to reduce non-specific interactions.
- Elution and Analysis: Elute ubiquitinated proteins with 2× SDS sample buffer containing 20 mM DTT at 95°C for 10 minutes. Analyze by Western blotting or mass spectrometry [4] [38].
Technological Workflows and Signaling Pathways
The following diagram illustrates the core workflow for UBD-based ubiquitin detection, highlighting critical steps where artifacts may be introduced and quality control measures should be implemented:
Diagram 1: UBD-based detection workflow.
The ubiquitin-proteasome system represents a complex signaling pathway that regulates protein fate within cells. The following diagram outlines key components and interactions in this system, highlighting points where detection methods intercept for analysis:
Diagram 2: Ubiquitin-proteasome system pathway.
Essential Research Reagent Solutions
The following table catalogues key reagents and materials essential for implementing robust ubiquitination detection assays while minimizing artifacts:
Table 2: Essential research reagents for ubiquitination studies
| Reagent/Material | Function | Application Notes |
|---|---|---|
| ThUBD Fusion Protein | High-affinity, unbiased capture of ubiquitin chains | Superior to TUBEs for sensitivity and linkage coverage [10] |
| Linkage-Specific Antibodies | Detection of specific ubiquitin chain types | Inherent linkage bias; K48 and K63 antibodies most common [4] |
| Anti-Ubiquitin Antibodies (P4D1, FK1, FK2) | General ubiquitin detection | Variable affinity; may not recognize all chain types equally [4] |
| Protein A/G Agarose Beads | Antibody immobilization for IP | Standard solid support for immunoprecipitation [38] |
| Protease Inhibitor Cocktails | Prevent protein degradation during processing | Essential for preserving ubiquitination signals [53] |
| N-Ethylmaleimide (NEM) | Deubiquitinase (DUB) inhibitor | Critical for maintaining ubiquitination status [4] |
| HDAC Inhibitors (e.g., TSA) | Prevent removal of ubiquitin-like modifiers | Important for studying related modifications |
| Cross-linking Reagents | Stabilize transient protein interactions | Can introduce artifacts if not optimized |
| High-Capacity 96-Well Plates | Platform for high-throughput assays | Corning 3603 type recommended for ThUBD coating [10] |
| Magnetic Beads with Streptavidin | Alternative solid support | Enable rapid separation; potential biotin interference [51] |
The objective comparison presented in this guide demonstrates that UBD-based methodologies, particularly those employing advanced ThUBD technology, offer significant advantages over traditional antibody-based approaches for comprehensive ubiquitination analysis in complex samples. The 16-fold enhancement in sensitivity and unbiased linkage recognition position ThUBD platforms as superior tools for researchers requiring accurate ubiquitination profiling, especially in drug discovery applications such as PROTAC development [10].
Antibody-based methods retain utility for targeted investigations of specific ubiquitin chain types, particularly when well-validated, high-affinity antibodies are available [4]. However, researchers must remain cognizant of the inherent limitations of linkage bias and potential for non-specific binding when interpreting results from these methods. The implementation of appropriate controls, including reference surfaces and blocking strategies, is essential for mitigating artifacts regardless of the chosen methodology [50] [51].
Future methodological developments will likely focus on further enhancing affinity and specificity of ubiquitin capture reagents, expanding high-throughput capabilities, and improving compatibility with emerging analytical technologies. As the ubiquitin field continues to evolve, researchers must carefully select detection methodologies that align with their specific experimental requirements while implementing robust controls to ensure the accurate interpretation of ubiquitination dynamics in health and disease.
Benchmarking Success: A Framework for Rigorous Method Comparison
In the study of protein ubiquitination, the efficiency of enrichment methodologies directly determines the accuracy and reliability of downstream analyses. Researchers primarily rely on two classes of tools for capturing ubiquitinated proteins: antibody-based reagents and ubiquitin-binding domain (UBD)-based technologies. The choice between these methods impacts every key metric—sensitivity, specificity, and precision—in profiling the ubiquitin code. This guide provides an objective, data-driven comparison of these approaches, focusing on their performance in experimental settings to inform method selection for basic research and drug development, particularly in areas such as PROTAC development where dynamic monitoring of ubiquitination is crucial [10].
Performance Metrics Comparison: UBD-Based vs. Antibody-Based Enrichment
The following table summarizes the quantitative performance data and characteristics of the leading enrichment methods.
| Metric/Method | TUBE-Based Platforms | ThUBD-Coated Plates | Linkage-Specific Antibodies |
|---|---|---|---|
| Detection Sensitivity | Baseline | 16-fold higher than TUBE [10] | Varies by antibody clone (e.g., FK2, P4D1) [4] |
| Dynamic Range | Limited capacity for polyUb chains [10] | 16-fold wider linear range than TUBE [10] | Not quantitatively specified |
| Linkage Bias | Yes, inherent bias towards certain chain types [10] [54] | Unbiased capture of all ubiquitin chain types [10] | Yes; linkage-specific (e.g., K48, K63) or pan-specific with varying affinity [4] |
| Key Advantage | High-throughput commercial availability | High-throughput, high-affinity, and unbiased capture | Ability to probe specific chain linkages from native tissues [4] |
| Primary Limitation | Low affinity and linkage bias can distort true ubiquitination status [10] | Requires production of specialized ThUBD fusion protein | High cost; potential for non-specific binding; cannot detect all chains simultaneously [4] |
Detailed Experimental Protocols and Methodologies
Understanding the experimental workflows is essential for interpreting the performance data and selecting the appropriate protocol.
Protocol 1: High-Throughput Ubiquitination Detection with ThUBD-Coated Plates
This protocol leverages a Tandem Hybrid Ubiquitin Binding Domain (ThUBD) for high-performance capture [10].
- Plate Coating: Coat Corning 3603 96-well plates with 1.03 μg ± 0.002 of the ThUBD fusion protein. The specific high-capacity plate type is critical for optimal performance [10].
- Sample Preparation and Incubation: Lyse cells or tissues under nondenaturing conditions to preserve ubiquitin chains. Add the complex proteome sample (with protein amounts within the validated dynamic range, e.g., as low as 0.625 μg) to the coated wells and incubate to allow ubiquitinated proteins to bind the ThUBD [10].
- Washing: Wash the plates with an optimized buffer to remove non-specifically bound proteins. The composition of this washing buffer is a key parameter determined during system optimization [10].
- Detection: Detect the captured ubiquitinated proteins using target-specific primary antibodies (for target-specific ubiquitination) or anti-ubiquitin antibodies (for global ubiquitination profiling). Quantification is performed with an HRP-conjugated secondary antibody and a chemiluminescent or colorimetric substrate [10].
Protocol 2: Enrichment Using TUBEs and Middle-Down Mass Spectrometry (UbiChEM-MS)
This protocol is used for enriching ubiquitin chains for detailed characterization via mass spectrometry, and is particularly useful for detecting branched chains [54].
- Enrichment: Incubate cell lysate (e.g., 45 mg from HEK293T cells) with agarose resin conjugated to Tandem Ubiquitin Binding Entities (TUBEs) overnight at 4°C [54].
- Washing: Pellet the resin and wash it with binding buffer followed by a minimal buffer (e.g., 50 mM Tris, 150 mM NaCl, pH 7.5) to remove contaminants [54].
- On-Resin Minimal Trypsinolysis: Digest the captured ubiquitin chains on-resin with trypsin at room temperature for 16 hours. The trypsin-to-lysate ratio must be empirically determined. This mild digestion cleaves ubiquitin only at Arg74, generating a Ub1–74 fragment [54].
- MS Analysis: Acidify the sample to stop the reaction, then analyze the supernatant by high-resolution mass spectrometry (e.g., Orbitrap Fusion Tribrid). A Ub1–74 fragment with a single Gly-Gly modification (Δ~114 Da) indicates a linear chain, while a fragment with two Gly-Gly modifications (Δ~228 Da) indicates a branch point within the ubiquitin chain [54].
Protocol 3: Immunoprecipitation with Ubiquitin Antibodies
This is a conventional method for enriching ubiquitinated proteins or specific chain types from native samples without genetic manipulation [55] [4].
- Antibody Immobilization: Incubate a high-affinity anti-ubiquitin antibody (e.g., pan-specific FK2 or linkage-specific K48 antibody) with Protein A/G agarose beads.
- Immunoprecipitation: Add the cell or tissue lysate to the antibody-bound beads and incubate to allow ubiquitinated proteins to bind.
- Washing: Pellet the beads and wash thoroughly with lysis buffer to remove non-specifically bound proteins.
- Elution and Analysis: Elute the enriched ubiquitinated proteins using low-pH buffer or Laemmli loading buffer. The eluate can then be analyzed by immunoblotting or mass spectrometry for downstream identification and quantification [4].
Visualizing the Ubiquitin Enrichment Workflows
The diagrams below illustrate the logical flow of the three core experimental protocols.
ThUBD Plate Assay Workflow
UbiChEM-MS Workflow
Antibody IP Workflow
The Scientist's Toolkit: Key Research Reagent Solutions
The following table details essential reagents and their functions for ubiquitination enrichment studies.
| Research Reagent | Function in Enrichment | Key Characteristics |
|---|---|---|
| ThUBD Fusion Protein | High-affinity, unbiased capture of all ubiquitin chain types in plate-based assays [10]. | Recombinant protein; core of a high-sensitivity platform. |
| TUBE (Tandem Ubiquitin Binding Entity) | Affinity purification of polyubiquitinated proteins from lysates [54] [55]. | Tandem UBDs increase affinity; can exhibit linkage bias. |
| Halo-NZF1 Resin | Selective enrichment of K29-linked ubiquitin chains for linkage-specific studies [54]. | Linkage-specific UBD (from TRABID DUB). |
| Anti-Ubiquitin Antibodies (P4D1, FK1/2) | Immunoprecipitation or blot detection of ubiquitinated proteins [54] [4]. | Pan-specific antibodies; FK2 prefers polyUb. |
| Linkage-Specific Ub Antibodies (e.g., α-K48) | Immunoprecipitation or blot detection of specific chain linkages [4]. | Essential for probing functions of specific Ub signals. |
| His-/Strep-Tagged Ubiquitin | Expression in cells to allow enrichment of ubiquitinated proteins via tag affinity [4]. | Genetic tool for proteomic-scale identification of substrates. |
In the evolving field of ubiquitinome research, the precision of enrichment methodologies critically determines the depth and accuracy of biological insights. Two principal approaches have emerged for capturing ubiquitinated proteins: antibody-based methods, which represent the traditional standard, and Ubiquitin-Binding Domain (UBD)-based strategies, which offer a more recent alternative. Antibody-based enrichment relies on immunoprecipitation using antibodies specific to ubiquitin or diglycine remnants, providing high specificity but facing challenges related to epitope accessibility and the dynamic nature of ubiquitination. In contrast, UBD-based methods utilize natural or engineered protein domains that physically interact with ubiquitin moieties, potentially offering broader recognition of diverse ubiquitin chain topologies. This comparison guide objectively evaluates the performance characteristics of these competing methodologies within the broader thesis that enrichment efficiency fundamentally dictates the quality of ubiquitinome data, influencing downstream biological interpretations and therapeutic development. The assessment is particularly relevant for researchers investigating complex biological processes such as cancer progression, immune regulation, and metabolic reprogramming where ubiquitination plays a pivotal role.
Performance Comparison: Quantitative Metrics
The following tables synthesize direct and indirect performance metrics for UBD-based and antibody-based enrichment methods from available experimental data.
Table 1: Direct Performance Metrics of UBD-based vs. Antibody-based Enrichment
| Performance Metric | UBD-based Method (DRUSP-ThUBD) | Traditional Antibody-based Method |
|---|---|---|
| Ubiquitin Signal Strength | Approximately 3 times stronger than control methods [24] | Baseline (Reference) |
| Overall Enrichment Efficiency | ~10-fold improvement in ubiquitin signal enrichment [24] | Baseline (Reference) |
| Enrichment Specificity | High (captures native ubiquitin spatial structures) [24] | High (dependent on antibody specificity) |
| Handling of Ubiquitin Chain Types | Efficient recognition of eight ubiquitin chain types without bias [24] | Variable (dependent on antibody specificity for chain types) |
Table 2: Practical and Experimental Considerations
| Consideration | UBD-based Method | Antibody-based Method |
|---|---|---|
| Sample Lysis Condition | Denatured-refolded (DRUSP) protocol minimizes DUB activity [24] | Typically native conditions, risk of DUB/proteasome activity [24] |
| Protein Extraction Efficiency | High (uses strongly denatured buffers) [24] | Potentially compromised (native lysis buffers) [24] |
| Reproducibility | Enhanced stability and reproducibility [24] | Can be undermined by contaminant proteins and DUBs [24] |
| Versatility | Compatible with various UBD types (pan-chains or chain-specific) [24] | Limited to available antibody repertoires |
| Key Application Evidence | Deep ubiquitinome profiling of mouse liver fibrosis [24] | Ubiquitin detection in IHC (e.g., human intestinal cancer tissue) [56] |
Experimental Protocols and Methodologies
The DRUSP-ThUBD Protocol for UBD-based Enrichment
The Denatured-Refolded Ubiquitinated Sample Preparation (DRUSP) method combined with a tandem hybrid UBD (ThUBD) represents a significant technical advancement. The protocol is designed to overcome key limitations of native lysis conditions and proceeds as follows [24]:
- Step 1: Denaturing Extraction - Tissue or cell samples are effectively extracted using strongly denatured buffers. This critical first step ensures sufficient protein extraction while simultaneously inactivating deubiquitinating enzymes (DUBs) and proteasomes that would otherwise remove ubiquitin signals during processing.
- Step 2: Filter-based Refolding - The denatured samples are subsequently refolded using filters to restore the native spatial structures of ubiquitin and ubiquitin chains, making them recognizable by the UBDs.
- Step 3: Tandem UBD Enrichment - The refolded samples are incubated with the engineered ThUBD, which efficiently captures the ubiquitinated proteins without bias toward different ubiquitin chain types.
- Step 4: Proteomic Analysis - The enriched ubiquitinated proteins are then identified and quantified using mass spectrometry, enabling comprehensive ubiquitinome profiling.
This workflow's effectiveness is evidenced by its application in deep ubiquitinome profiling of early mouse liver fibrosis, which revealed novel insights with increased accuracy [24].
Standard Antibody-based Immunoprecipitation Protocol
Traditional antibody-based methods for ubiquitin enrichment typically follow this general workflow [24] [56]:
- Step 1: Native Lysis - Cells or tissues are lysed under native conditions using mild, non-denaturing buffers to preserve protein-protein interactions and ubiquitin epitopes.
- Step 2: Antibody Incubation - The lysate is incubated with antibodies specific to ubiquitin, diglycine remnants (K-ε-GG), or particular ubiquitin chain types.
- Step 3: Capture and Washing - The antibody-antigen complexes are captured using protein A/G beads, followed by extensive washing to remove non-specifically bound proteins.
- Step 4: Elution and Analysis - Bound ubiquitinated proteins are eluted, typically under denaturing conditions, and prepared for downstream mass spectrometry analysis.
A key limitation of this approach is the use of native lysis conditions, which presents significant challenges including insufficient protein extraction, heightened activity of DUBs and proteasomes in removing ubiquitin signals, and co-purification of contaminant proteins [24].
Biological Context and Signaling Pathways
Understanding the biological significance of ubiquitination highlights the importance of efficient enrichment methods. UBD (Ubiquitin D), also known as FAT10, is not just a tool but a biologically significant ubiquitin-like modifier itself. It plays crucial roles in cancer progression through multiple mechanisms:
Figure 1: UBD's Role in Cancer Progression and Immune Evasion
In colorectal cancer, UBD promotes cell proliferation by facilitating the degradation of the tumor suppressor p53 [57]. UBD interacts with p53 and downregulates its expression by regulating its degradation through the proteasome system, shortening the p53 half-life. This subsequently affects key cell cycle regulators, leading to decreased p21 and increased Cyclin D1, Cyclin E, and various CDKs [57].
Simultaneously, in the tumor microenvironment of cancers like ovarian cancer, UBD-mediated glycolytic reprogramming promotes M2 macrophage polarization [58]. UBD upregulation correlates with increased expression of key glycolytic enzymes such as LDHA and ALDOA. This metabolic shift enhances M2 macrophage polarization, which in turn facilitates immune evasion and immunotherapy resistance [58].
Figure 2: Technical Workflow Comparison for Ubiquitin Enrichment
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for UBD and Ubiquitin Research
| Reagent / Tool | Function / Application | Examples / Specifications |
|---|---|---|
| Tandem Hybrid UBD (ThUBD) | High-efficiency enrichment of ubiquitinated proteins; recognizes multiple chain types without bias [24] | Engineered construct for DRUSP protocol |
| UBD/FAT10 Antibodies | Detection of UBD expression in cells and tissues via IHC, WB [57] [56] | Polyclonal Antibody (PA5-80201); validated for IHC in human cancer tissue [56] |
| Denatured-Refolded Sample Preparation (DRUSP) | Protocol to maximize ubiquitin signal preservation by minimizing DUB activity [24] | Uses strong denaturing buffers followed by filter-based refolding |
| Proteasome Inhibitors | Block degradation of ubiquitinated proteins; essential for preserving ubiquitin signals | MG132 [57] |
| UBD Expression Plasmids | Functional studies of UBD effects in cell models | Lentiviral vectors for stable expression [57] |
| Mass Spectrometry Platforms | Identification and quantification of enriched ubiquitinated proteins | High-resolution LC-MS/MS systems |
The head-to-head comparison between UBD-based and antibody-based enrichment methods reveals a nuanced performance landscape. The DRUSP-ThUBD methodology demonstrates superior quantitative performance in both ubiquitin signal strength (approximately 3× stronger) and overall enrichment efficiency (~10× improvement) compared to traditional antibody-based approaches [24]. Furthermore, its capacity to work under denaturing conditions that minimize DUB activity and its unbiased recognition of diverse ubiquitin chain types represent significant technical advantages.
However, antibody-based methods maintain relevance for specific applications, particularly immunohistochemical analysis of tissue samples where spatial localization is essential [57] [56]. The choice between methodologies should be guided by research priorities: UBD-based methods for comprehensive ubiquitinome profiling where depth and accuracy are paramount, and antibody-based approaches for targeted studies requiring cellular localization or validation of specific ubiquitination events.
For the field of drug development, particularly in cancer therapeutics where ubiquitination pathways are increasingly targeted, the enhanced reproducibility and quantitative accuracy of UBD-based methods offer more reliable platforms for biomarker discovery and mechanistic studies. As research continues to unravel the complexity of the ubiquitin code, methodological advances that improve our capacity to capture this diversity will directly translate to accelerated therapeutic development.
Ubiquitination is a critical post-translational modification (PTM) that regulates diverse cellular functions, including protein degradation, signal transduction, and DNA repair [4]. The versatility of ubiquitin signaling stems from the complexity of ubiquitin conjugates, which can form various chain architectures with different linkage types [4]. While K48- and K63-linked chains are well-characterized, atypical ubiquitin chains (K6, K11, K27, K29, K33, and M1-linked) present significant detection challenges due to their lower abundance and the historical lack of specific tools for their enrichment and identification [4].
This case study analysis objectively compares the performance of Ubiquitin Binding Domain (UBD)-based methods versus antibody-based approaches for enriching and detecting atypical ubiquitin chains. We evaluate these methodologies based on affinity, specificity, throughput, and applicability to drug discovery pipelines, with a particular focus on their utility in profiling branched and atypical chain architectures that are increasingly recognized as biologically significant [59].
Fundamental Principles of Ubiquitin Chain Recognition
Both UBD-based and antibody-based methods aim to overcome the central challenge in ubiquitinomics: the low stoichiometry of ubiquitinated substrates within complex proteomes [4]. These approaches facilitate the enrichment of ubiquitinated proteins or peptides prior to analysis by western blotting or mass spectrometry.
- UBD-Based Methods: Utilize natural ubiquitin-binding domains engineered for enhanced affinity and specificity. These include Tandem Ubiquitin Binding Entities (TUBEs) and the more recently developed Tandem Hybrid Ubiquitin Binding Domain (ThUBD) [10] [60].
- Antibody-Based Methods: Employ antibodies that recognize ubiquitin epitopes. Pan-specific antibodies (e.g., P4D1, FK1/FK2) recognize all ubiquitin linkages, while linkage-specific antibodies target particular chain types (e.g., K11-, K27-, K48-, or K63-linkages) [4].
Key Technological Platforms
The experimental workflows for UBD-based and antibody-based methods share common objectives but differ significantly in their implementation and underlying recognition mechanisms. The following diagram illustrates the core logical relationship between these methodological approaches and their applications:
Figure 1: Methodological Approaches for Ubiquitin Chain Analysis. This workflow outlines the two primary enrichment strategies for ubiquitinated proteins and their downstream applications.
Comparative Performance Analysis
Affinity and Sensitivity Benchmarks
A critical comparison of the detection capabilities between ThUBD-coated plates and TUBE-coated plates reveals significant performance differences, particularly relevant for detecting low-abundance atypical chains.
Table 1: Performance Comparison of UBD-Based Technologies
| Parameter | ThUBD-Coated Plates | TUBE-Coated Plates |
|---|---|---|
| Detection Sensitivity | As low as 0.625 μg | 16-fold lower sensitivity |
| Dynamic Range | 16-fold wider linear range | Limited linear range |
| Throughput Capacity | High-density 96-well format | High-density 96-well format |
| Ubiquitin Chain Bias | Unbiased recognition | Linkage-dependent bias |
| Affinity | High affinity for polyubiquitin chains | Lower affinity for ubiquitin chains |
The 16-fold enhancement in dynamic range and significantly improved detection sensitivity position ThUBD technology as particularly advantageous for capturing low-abundance atypical ubiquitin chains that are challenging to detect with conventional methods [10].
Linkage Recognition Specificity and Bias
The ability to accurately profile heterogeneous ubiquitin chain architectures depends heavily on the linkage recognition properties of the enrichment tool.
Antibody-Based Limitations: Traditional antibodies exhibit variable affinity for different linkage types, potentially leading to biased representation of chain populations. While linkage-specific antibodies are invaluable for confirming particular chain types, their application for comprehensive ubiquitinome profiling is limited by this inherent bias [4] [61].
UBD-Based Advantages: Engineered ThUBD combines multiple ubiquitin-binding domains to achieve unbiased recognition across all ubiquitin chain types while maintaining high affinity. This unbiased approach is essential for accurately capturing the complexity of atypical chain architectures, including heterotypic and branched chains [10].
Application in Functional Studies and Drug Discovery
The utility of ubiquitin detection methods extends beyond basic research into applied drug discovery, particularly in the development of Proteolysis-Targeting Chimeras (PROTACs).
Table 2: Application Scope in Research and Drug Development
| Application | Antibody-Based Methods | UBD-Based Methods |
|---|---|---|
| Global Ubiquitination Profiling | Limited by linkage bias | Suitable with ThUBD |
| Target-Specific Ubiquitination | Linkage-specific antibodies enable this | Chain-specific TUBEs enable this |
| PROTAC Development | Not ideal for high-throughput screening | Ideal for high-throughput assays |
| Endogenous Protein Analysis | Suitable for clinical samples | Suitable with genetic manipulation |
| Cellular Ubiquitination Dynamics | Limited by antibody availability | TUBEs can monitor dynamic changes |
Chain-specific TUBEs have demonstrated particular utility in discriminating K48-linked (PROTAC-mediated degradation) versus K63-linked (inflammatory signaling) ubiquitination events on endogenous proteins like RIPK2, highlighting their value in mechanism-of-action studies for targeted protein degradation therapeutics [60].
Experimental Protocols for Ubiquitin Chain Analysis
High-Throughput ThUBD-Coated Plate Protocol
The ThUBD-coated plate methodology represents a significant advancement for sensitive, high-throughput ubiquitination detection [10]:
- Plate Preparation: Coat Corning 3603-type 96-well plates with 1.03 μg ± 0.002 of ThUBD fusion protein.
- Sample Preparation: Lyse cells or tissues in ubiquitination-preserving lysis buffer (lacking EDTA and containing 1% NP-40, 50 mM Tris-HCl pH 7.5, 150 mM NaCl, and complete protease inhibitors).
- Capture Incubation: Add 5 pmol of polyubiquitin chains or complex proteome samples to ThUBD-coated wells and incubate for 2 hours at 4°C with gentle agitation.
- Washing: Remove non-specifically bound proteins with three washes using TBST buffer (Tris-buffered saline with 0.1% Tween-20).
- Detection: Incubate with primary antibody against target protein or ubiquitin, followed by HRP-conjugated secondary antibody. Develop with chemiluminescent substrate and quantify signals.
This protocol enables specific detection of ubiquitination signals from as little as 0.625 μg of proteome sample, making it suitable for limited clinical samples or rare cell populations [10].
Chain-Specific TUBE Assay for PROTAC Evaluation
For assessing linkage-specific ubiquitination in PROTAC development [60]:
- Cell Treatment: Treat THP-1 cells or other relevant cell lines with PROTAC compounds or molecular glues for predetermined timepoints.
- Cell Lysis: Harvest cells and lyse in specialized buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, 10% glycerol, 1 mM PMSF, and 10 mM N-ethylmaleimide to preserve ubiquitination).
- Affinity Capture: Incubate cell lysates with chain-specific TUBEs (K48-TUBE for PROTAC degradation, K63-TUBE for inflammatory signaling) immobilized on 96-well plates.
- Target Detection: Detect captured ubiquitinated targets using target-specific primary antibodies and HRP-conjugated secondary antibodies.
- Quantification: Normalize ubiquitination signals to total target protein levels and express as fold-change over vehicle control.
This approach successfully differentiated K48-linked ubiquitination induced by RIPK2 PROTAC from K63-linked ubiquitination stimulated by L18-MDP inflammatory stimulus, demonstrating the utility of chain-specific TUBEs in elucidating mechanism of action for ubiquitination-modulating therapeutics [60].
Biological Significance of Atypical Ubiquitin Chains
Structural Insights into Branched Ubiquitin Chain Recognition
Recent cryo-EM studies have revealed the biological importance of K11/K48-branched ubiquitin chains in proteasomal recognition and degradation. These structures constitute 10-20% of cellular ubiquitin polymers and function as priority degradation signals during cell cycle progression and proteotoxic stress [59].
The human 26S proteasome recognizes K11/K48-branched chains through a multivalent binding mechanism involving:
- A novel K11-linked ubiquitin binding site at the interface of RPN2 and RPN10
- The canonical K48-linkage binding site formed by RPN10 and RPT4/5
- RPN2 recognition of alternating K11-K48-linkages through a conserved motif [59]
This structural understanding explains why K11/K48-branched ubiquitin chains serve as enhanced proteasomal degradation signals compared to homotypic chains and underscores the importance of tools that can specifically detect these complex architectures.
Functional Roles in Cellular Signaling and Disease
Atypical ubiquitin chains regulate diverse cellular processes beyond proteasomal degradation:
- K63-linked chains regulate protein-protein interactions in the NF-κB pathway and autophagy [4]
- M1-linked linear chains activate inflammatory signaling pathways [4]
- K11/K48-branched chains mediate timely degradation of mitotic regulators and pathological Huntingtin variants [59]
- UBD (Ubiquitin D) overexpression activates p38 MAPK signaling in rheumatoid arthritis, promoting fibroblast-like synoviocyte proliferation and inflammatory cytokine secretion [62]
The association of atypical ubiquitin chains with specific disease pathologies highlights the therapeutic relevance of methods that can accurately detect these modifications in physiological contexts.
Research Reagent Solutions
Table 3: Essential Research Reagents for Ubiquitin Enrichment Studies
| Reagent/Tool | Function | Application Context |
|---|---|---|
| ThUBD-Coated Plates | Unbiased, high-affinity capture of all ubiquitin chain types | High-throughput global ubiquitination profiling |
| Chain-Specific TUBEs | Selective enrichment of particular linkage types | Differentiating K48 vs. K63 ubiquitination in PROTAC screening |
| Linkage-Specific Antibodies | Immunodetection of specific ubiquitin linkages | Validation of particular chain types by western blot |
| His/Strep-Tagged Ubiquitin | Affinity purification of ubiquitinated substrates | Identification of ubiquitination sites by mass spectrometry |
| DUB Inhibitors (N-ethylmaleimide) | Preserve ubiquitination signatures during processing | Maintain ubiquitin chain integrity in cell lysates |
| Size-Exclusion Chromatography | Enrich medium-length ubiquitin chains (n=4-8) | Preparation of defined ubiquitin chains for structural studies |
This case study analysis demonstrates that both UBD-based and antibody-based methods offer distinct advantages for detecting atypical ubiquitin chains, with the optimal choice depending on the specific research objectives.
- ThUBD-based platforms provide superior sensitivity and unbiased recognition for comprehensive ubiquitinome profiling, making them ideal for discovery-phase research where the complete spectrum of ubiquitin linkages needs to be captured.
- Chain-specific TUBEs offer the unique capability to differentiate linkage-specific ubiquitination events on endogenous proteins, providing valuable mechanistic insights in PROTAC development and signaling studies.
- Antibody-based methods remain valuable for targeted validation of specific ubiquitin chain types, particularly in clinical samples where genetic manipulation is not feasible.
The continued development of enhanced ubiquitin detection methodologies will be essential for unraveling the complex biological functions of atypical ubiquitin chains and harnessing this knowledge for therapeutic innovation in areas ranging from targeted protein degradation to inflammatory disease treatment.
Ubiquitination, the covalent attachment of ubiquitin to substrate proteins, is a crucial post-translational modification regulating diverse cellular processes including protein degradation, cell signaling, and immune response [37] [4]. The scientific exploration of the "ubiquitinome" – the complete set of ubiquitinated proteins in a biological system – faces significant technical challenges due to the low abundance of ubiquitinated substrates (less than 1% of the total proteome), the complexity of ubiquitin chain architectures, and the dynamic nature of ubiquitination signals which are constantly removed by deubiquitinating enzymes (DUBs) [37] [4]. To overcome these challenges, researchers have developed sophisticated enrichment methodologies, primarily falling into two categories: ubiquitin-binding domain (UBD)-based approaches and antibody-based immunotechnologies. This guide provides a comprehensive comparative analysis of the emerging tandem hybrid UBD (ThUBD) platform against established high-affinity antibody methods, evaluating their performance characteristics, experimental requirements, and suitability for different research scenarios. The evaluation is framed within the critical context of enrichment efficiency – a paramount consideration for deep and accurate ubiquitinome profiling.
Tandem Hybrid Ubiquitin-Binding Domains (ThUBDs)
ThUBDs represent an engineered protein-based approach for ubiquitinated protein capture. These synthetic reagents are constructed by systematically combining multiple natural ubiquitin-binding domains (UBDs) with high affinity for different ubiquitin configurations into a single tandem hybrid molecule [11]. The design process involves selecting UBDs with high affinity to different types of ubiquitin chains, then evaluating various UBD combinations with different lengths and types to create optimized constructs [11]. Examples include ThUDQ2 (combining DSK2p-derived UBA and ubiquilin 2-derived UBA domains) and ThUDA20 (combining DSK2p-derived UBA and RABGEF1-derived A20-ZnF domains) [11]. The fundamental operating principle relies on the natural affinity between UBDs and the hydrophobic surfaces of ubiquitin molecules (such as Ile44 and Ile36), which serve as the structural basis for recognition [37]. This design achieves markedly higher affinity for ubiquitinated proteins compared to naturally occurring single UBDs and displays almost unbiased high affinity to all seven lysine-linked ubiquitin chains [11].
High-Affinity Antibody-Based Methods
Antibody-based approaches leverage the specific binding between antibodies and ubiquitin epitopes. These methods primarily utilize two strategies: (1) antibodies that recognize all ubiquitin linkages (such as P4D1, FK1/FK2), and (2) linkage-specific antibodies that target particular ubiquitin chain architectures (M1-, K11-, K27-, K48-, or K63-linkage specific antibodies) [4]. The antibodies function by recognizing specific linear epitopes or spatial structures on ubiquitin or ubiquitin chains. For research requiring tagged ubiquitin systems, epitope tags (Flag, HA, V5, Myc, Strep, His) can be employed, with subsequent enrichment using tag-specific antibodies or resins [4]. Antibody-based purification methods range from crude physicochemical fractionation to highly specific antigen-affinity approaches, with class-specific affinity purification using bacterial proteins (Protein A, G, L) being particularly common for antibody isolation itself [63].
Performance Comparison: Quantitative Data Analysis
The following table summarizes key performance metrics for ThUBD and antibody-based methods based on recent experimental findings:
Table 1: Performance Comparison of Ubiquitin Enrichment Methods
| Performance Metric | ThUBD-Based Approach | Traditional Antibody-Based Methods | Experimental Context |
|---|---|---|---|
| Overall Ubiquitin Signal Enrichment | ~10-fold improvement over control [37] | Not quantified (varies by antibody) | Comparison to non-enriched controls |
| Ubiquitin Signal Intensity | ~3x stronger than native control [37] | Baseline reference | DRUSP method with ThUBD vs. native methods |
| Linkage Recognition Profile | Unbiased high affinity to 8 ubiquitin chains [37] [11] | Varies by antibody; some linkage-specific | Pan-specific vs. linkage-specific applications |
| Identified Ubiquitinated Proteins | 7,487 mammalian proteins [11] | 96-753 proteins [4] | Mammalian cells (varied by study) |
| Structural Dependency | Requires native ubiquitin structure [37] | Depends on antibody (linear vs. conformational) | Recognition constraints |
| Typical Contaminant Levels | Reduced co-purification [37] | Non-specific binding observed [4] | Specificity in complex lysates |
Experimental Protocols: Methodological Considerations
ThUBD Workflow: DRUSP Protocol
The Denatured-Refolded Ubiquitinated Sample Preparation (DRUSP) protocol combined with ThUBD enrichment represents a recent methodological advancement [37]:
- Sample Extraction and Denaturation: Tissues or cells are lysed using strong denaturing buffers (e.g., 4% SDS) to effectively inactivate DUBs and proteasomes, while simultaneously improving the extraction efficiency of insoluble ubiquitinated proteins. A cryogenic freeze grinder can be utilized for tissue disruption.
- Refolding Step: The denatured lysate is subjected to a buffer exchange using filters to remove denaturants and allow refolding of ubiquitin and ubiquitin chains into their native spatial structures, which is essential for ThUBD recognition.
- ThUBD Enrichment: The refolded sample is incubated with immobilized ThUBD. The tandem hybrid design, incorporating multiple high-affinity UBDs, enables efficient capture of ubiquitinated proteins with minimal linkage bias.
- Wash and Elution: After extensive washing to remove non-specifically bound proteins, the enriched ubiquitinated proteins are eluted, typically using acidic buffers or SDS-PAGE loading buffer.
- Downstream Analysis: The eluate is digested with trypsin and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) for ubiquitinome profiling [37].
Antibody-Based Immunoaffinity Protocol
Standard protocols for antibody-based ubiquitin enrichment typically follow these steps:
- Cell Lysis under Native Conditions: Cells are lysed using nondenaturing buffers to preserve the ubiquitin epitopes recognized by the antibodies. Protease and DUB inhibitors are commonly added, though their efficacy can be incomplete under these conditions [37].
- Antibody Immobilization: The capture antibody (e.g., FK2 for total ubiquitin, or linkage-specific antibodies) is coupled to solid support beads, such as agarose or magnetic beads.
- Immunoaffinity Capture: The clarified lysate is incubated with the antibody-coupled beads to allow binding of ubiquitinated proteins.
- Stringent Washes: Beads are washed multiple times with nondenaturing buffers to reduce non-specific background proteins. The challenge of contaminant proteins co-purifying under native conditions is noted as a significant issue [37].
- Elution and Digestion: Bound ubiquitinated proteins are eluted, often with low-pH buffer or by competition with free ubiquitin peptide. For diGly remnant analysis (the signature of ubiquitinated peptides after trypsin digestion), the enriched proteins are digested with trypsin.
- Peptide-Level Enrichment (Optional): For increased specificity, a second enrichment step using anti-K-ε-GG antibodies to isolate peptides containing the diGly modification can be performed, followed by LC-MS/MS analysis [4].
Workflow Visualization
Diagram 1: Comparative experimental workflows for ubiquitin enrichment.
Advantages, Limitations, and Application Scenarios
Comparative Strengths and Weaknesses
Table 2: Strategic Comparison of Ubiquitin Enrichment Methods
| Feature | ThUBD Platform | Antibody-Based Platform |
|---|---|---|
| Key Advantages | Superior signal enrichment; Minimal linkage bias; Enhanced reproducibility; Effective for insoluble proteins (via DRUSP) [37] [11]. | Direct application to clinical/animal tissues; Linkage-specific analysis possible; Well-established protocols [4]. |
| Inherent Limitations | Requires refolded/native ubiquitin structure; Not suitable for fixed samples; Genetic manipulation not feasible in tissues [37]. | Potential high cost of quality antibodies; Non-specific binding; Epitope masking possible; Variable performance under native lysis [37] [4]. |
| Optimal Use Cases | Deep, comprehensive ubiquitinome profiling; Discovery-phase research; Studies requiring broad chain-type analysis [37]. | Targeted validation studies; Analysis of specific chain linkages; Pathological samples where genetic tagging is impossible [4]. |
Decision Framework for Researchers
Choosing between ThUBD and antibody-based methods depends on several research parameters:
- Research Objective: For hypothesis-generating, global ubiquitinome mapping, ThUBD offers clear advantages in depth and breadth. For validating specific ubiquitination events or studying particular chain types, well-validated linkage-specific antibodies may be preferable.
- Sample Type: For cell lines where genetic manipulation is feasible, ThUBD applied to DRUSP-prepared samples yields exceptional results. For archived clinical tissues or animal samples, antibody-based methods remain the only viable option.
- Technical Resources: ThUBD protocols require expertise in protein refolding and handling. Antibody-based methods are often more readily implemented in standard molecular biology laboratories.
- Budget Considerations: While recombinant ThUBD production has associated costs, the high price of high-quality antibodies (particularly linkage-specific ones) can also be significant.
Essential Research Reagent Solutions
The following table catalogs key reagents required for implementing the ubiquitin enrichment methodologies discussed in this guide.
Table 3: Key Research Reagents for Ubiquitin Enrichment Studies
| Reagent / Solution | Function / Purpose | Example Applications |
|---|---|---|
| Tandem Hybrid UBD (ThUBD) | High-affinity, unbiased capture of ubiquitinated proteins; Core enrichment reagent [37] [11]. | Ubiquitinome profiling under denatured-refolded (DRUSP) or native conditions. |
| Pan-Specific Ub Antibodies (e.g., P4D1, FK2) | Immunoaffinity enrichment of total ubiquitinated proteins; Recognizes various ubiquitin linkages [4]. | Western blot validation; Immunoprecipitation of ubiquitinated proteins from native lysates. |
| Linkage-Specific Ub Antibodies | Selective isolation of proteins modified with specific ubiquitin chain types (e.g., K48, K63) [4]. | Studying the biology of specific ubiquitin signals (e.g., proteasomal degradation vs. signaling). |
| Strong Denaturing Lysis Buffer (e.g., 4% SDS) | Efficiently inactivates DUBs and proteasomes; improves extraction of insoluble proteins [37]. | Critical first step in the DRUSP protocol to preserve the ubiquitinome. |
| Affinity Purification Resins | Solid support for immobilizing ThUBD or antibodies (e.g., glutathione-sepharose for GST-tagged ThUBD, protein A/G for antibodies) [63] [11]. | Performing the enrichment pull-down from complex biological lysates. |
| Protease & DUB Inhibitors | Protects ubiquitination signals from enzymatic degradation during sample preparation [37]. | Added to lysis buffers, especially in native protocols, to maintain modification integrity. |
The comparative analysis presented in this guide demonstrates that ThUBD-based methodologies, particularly when integrated with the DRUSP protocol, establish a new benchmark for enrichment efficiency in ubiquitinomics research. The quantitative data reveals a significant advantage in terms of ubiquitin signal strength, depth of proteome coverage, and methodological reproducibility [37]. Antibody-based approaches, while historically foundational and still essential for specific applications like linkage-specific analysis or work with intractable sample types, face challenges related to specificity, stability of ubiquitin signals under native conditions, and co-purification of contaminating proteins.
The future of ubiquitinome research will likely see further engineering of UBD-based capture reagents with enhanced affinities and specificities. Furthermore, the integration of artificial intelligence and machine learning for predicting antibody-antigen interactions and optimizing protein binders is already transforming the therapeutic antibody landscape and may soon influence research reagent development [64]. For the practicing researcher, the choice between these "gold standard" tools is not absolute but must be strategically aligned with the specific biological question, sample resources, and technical constraints. However, for applications demanding the most comprehensive and accurate picture of the ubiquitinome, ThUBD-based platforms currently represent the leading edge of technological capability.
For researchers profiling protein ubiquitination, selecting the appropriate enrichment method is crucial for experimental success. The choice between Ubiquitin D (UBD)-based approaches and antibody-based methods involves significant trade-offs in throughput, specificity, cost, and practical applicability. The following table summarizes the core characteristics of each method to guide your selection.
| Feature | UBD-Based Tagging Approaches | Antibody-Based Enrichment |
|---|---|---|
| Core Principle | Expression of affinity-tagged Ub (e.g., His, Strep) in cells; covalent labeling of substrates [4] | Immunoaffinity purification using anti-Ub antibodies (e.g., P4D1, FK2) or linkage-specific antibodies [4] |
| Primary Application | High-throughput screening of ubiquitinated substrates in cultured cells [4] | Profiling endogenous ubiquitination under physiological conditions; linkage-specific analysis [4] |
| Throughput | High (amenable to proteomic-scale screening) [4] | Moderate (lower throughput due to enrichment steps) [4] |
| Specificity & Key Advantage | Easy, low-cost friendly method for substrate screening [4] | Recognizes endogenous ubiquitination; enables research on specific chain linkages (e.g., K48, K63) [4] |
| Major Limitation | Potential for artifacts; not feasible for animal or patient tissues [4] | High cost of antibodies; potential for non-specific binding [4] |
| Identification Efficiency | Relatively low [4] | Higher for endogenous proteins; can be applied to tissue samples [4] |
| Cost Consideration | Lower upfront cost; requires genetic cell line construction [4] | Higher reagent cost; no need for genetic manipulation [4] |
Detailed Experimental Protocols
Protocol for UBD Tagging-Based Enrichment
This protocol is designed for the large-scale identification of ubiquitination sites in cultured cells [4].
- Step 1: Cell Line Engineering
- Construct a cell line (e.g., HEK293T, U2OS) that stably expresses ubiquitin (Ub) tagged with an affinity tag, such as 6x-His or Strep-tag [4].
- Step 2: Protein Harvest and Lysis
- Grow the engineered cells under standard conditions.
- Harvest cells and lyse them using a denaturing buffer (e.g., containing 6 M Guanidine-HCl) to preserve the ubiquitination state and inactivate deubiquitinases.
- Step 3: Affinity Purification
- Step 4: On-Bead Digestion
- Subject the purified, ubiquitinated proteins to on-bead tryptic digestion.
- Step 5: Mass Spectrometry (MS) Analysis
- Analyze the resulting peptides by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
- Identify ubiquitination sites by searching for a diagnostic mass shift of 114.04 Da on modified lysine residues [4].
Protocol for Antibody-Based Enrichment
This method is optimal for studying endogenous ubiquitination or specific Ub chain linkages in cell lines or patient-derived tissues [4].
- Step 1: Sample Preparation
- Lyse cells or homogenize tissue samples in a suitable lysis buffer. The use of a denaturing buffer is recommended to maintain the integrity of the ubiquitinome.
- Step 2: Immunoaffinity Enrichment
- Step 3: Stringent Washing
- Wash the resin extensively to remove non-specifically bound proteins.
- Step 4: Elution and Digestion
- Elute the bound ubiquitinated proteins using a low-pH buffer or by directly denaturing the resin in an SDS-containing buffer.
- Digest the eluted proteins into peptides for MS analysis.
- Step 5: LC-MS/MS and Data Analysis
- Perform LC-MS/MS analysis and database searching to identify ubiquitinated peptides and their modification sites.
Diagram 1: Experimental selection workflow for ubiquitination studies.
Cost-Benefit and Operational Analysis
Quantitative Cost & Performance Comparison
A holistic decision requires balancing scientific needs with project constraints. The following table provides a comparative analysis of key operational factors.
| Aspect | UBD-Based Tagging | Antibody-Based Enrichment |
|---|---|---|
| Relative Cost | Lower reagent cost [4] | High (cost of specific antibodies) [4] |
| Handling of Endogenous Proteins | No (requires genetic manipulation) [4] | Yes [4] |
| Tissue Sample Compatibility | Not feasible [4] | Yes, directly applicable [4] |
| Sample Throughput | High (once stable line is established) [4] | Moderate (per-sample enrichment needed) [4] |
| Lead Time | Long (weeks for cell line generation) | Short (days for protocol setup) |
| Data Specificity | Provides global ubiquitinome profile [4] | Can be global or linkage-specific [4] |
| Risk of Artifacts | Higher (tagged Ub may not fully mimic endogenous Ub) [4] | Lower (recognizes natural proteins) |
Simulation and Optimization in Decision-Making
Given the high cost and complexity of wet-lab experiments, simulation and optimization modeling can be valuable decision-support tools. A well-constructed model can help justify the investment in a particular methodological path.
- Stochastic Cost-Benefit Analysis (CBA): CBA quantifies and compares expected costs and benefits over time. Incorporating stochastic (variable) parameters provides a more realistic financial assessment. For instance, simulation can model variable factors like success rates, identification yields, and resource consumption to estimate the economic landscape of a digitalisation project, which in a research context translates to adopting a new platform technology [65].
- Linear Programming for Resource Optimization: Linear programming algorithms can be employed to simulate behavior and derive optimal solutions, such as cost minimization or resource maximization. This is particularly powerful for handling large datasets and linear relationships, which can inform efficient resource allocation in large-scale proteomic projects [66].
- Practical CBA for Projects: A practical cost-benefit analysis for an optimization project involves observing current processes, interviewing personnel, and analyzing historical data to build a baseline. Creating different scenarios (conservative, moderate, aggressive) helps estimate potential benefits, Return on Investment (ROI), and Time To Payback (TTP), which can be presented via custom templates for clarity [67].
Diagram 2: Cost-benefit analysis and simulation workflow for project planning.
The Scientist's Toolkit: Key Research Reagent Solutions
Selecting the right reagents is fundamental for implementing either enrichment protocol. The table below details essential materials and their functions.
| Reagent / Material | Function in Experiment |
|---|---|
| Affinity-Tagged Ubiquitin (e.g., 6x-His-Ub, Strep-Ub) | Engineered ubiquitin used in UBD-based methods to covalently label cellular substrates for subsequent purification [4]. |
| Anti-Ubiquitin Antibodies (e.g., P4D1, FK2) | Immunoaffinity reagents for pulldown of endogenously ubiquitinated proteins from complex lysates [4]. |
| Linkage-Specific Anti-Ub Antibodies (e.g., α-K48, α-K63) | Antibodies that recognize specific ubiquitin chain linkages, enabling the study of the unique signals conferred by different chain types [4]. |
| Ni-NTA Agarose Resin | Chromatographic resin that binds with high affinity to polyhistidine (6x-His) tags, used for purifying His-tagged ubiquitin conjugates [4]. |
| Strep-Tactin Resin | Affinity resin with high specificity and affinity for Strep-tag II, used for purifying Strep-tagged ubiquitin conjugates under gentle conditions [4]. |
| Deubiquitinase (DUB) Inhibitors | Added to lysis buffers to prevent the cleavage of ubiquitin from substrates by endogenous DUBs, thereby preserving the ubiquitinome profile. |
| Trypsin | Protease used to digest purified proteins into peptides for downstream analysis by mass spectrometry. |
The choice between UBD-based and antibody-based enrichment is not a matter of which method is universally superior, but which is most appropriate for your specific research question and constraints.
- For foundational, discovery-phase research in tractable cell lines, where cost-effectiveness and high-throughput capability are priorities, UBD-based tagging offers a powerful and efficient starting point [4].
- For clinically-oriented research, studies requiring analysis of endogenous regulation, or investigations into specific Ub chain signaling, the antibody-based approach is indispensable, despite its higher cost, due to its direct applicability to tissues and superior specificity [4].
Employing computational simulations and cost-benefit analyses as part of the project planning stage can significantly de-risk this decision, providing a data-driven framework for selecting the most efficient and economically viable path for your research on protein ubiquitination [65] [67].
Conclusion
The choice between UBD-based and antibody-based enrichment is not one-size-fits-all but should be guided by the specific research objectives. UBDs, particularly engineered tandem domains like ThUBD, offer superior, unbiased affinity for diverse ubiquitin chains and are ideal for high-throughput, sensitive profiling of the global ubiquitinome. In contrast, antibody-based methods provide a direct, often linkage-specific approach suitable for validating targets or studying particular chain types, though they can be limited by affinity and bias. Future directions will be shaped by the continued engineering of high-affinity binders, the integration of artificial intelligence for method optimization, and the development of novel platforms that merge the strengths of both approaches. Ultimately, a critical and comparative understanding of these tools is fundamental for advancing our knowledge of ubiquitin signaling and translating these insights into novel therapeutics for cancer, neurodegenerative diseases, and beyond.
References
- 1. The role of ubiquitination in health and disease [https://pubmed.ncbi.nlm.nih.gov/39329019/]
- 2. Emerging functions of branched ubiquitin chains [https://www.nature.com/articles/s41421-020-00237-y]
- 3. Visualizing ubiquitination in mammalian cells - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6362358/]
- 4. Current methodologies in protein ubiquitination ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9373315/]
- 5. Data-independent acquisition method for ubiquitinome ... [https://www.nature.com/articles/s41467-020-20509-1]
- 6. Multiepitope recognition technology promotes the in-depth ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11628813/]
- 7. A versatile new tool derived from a bacterial... | PLOS Biology [https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3001501]
- 8. The Ubiquitin Tale: Current Strategies and Future Challenges [https://pmc.ncbi.nlm.nih.gov/articles/PMC11406696/]
- 9. The Molecular Toolbox for Linkage Type‐Specific Analysis ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12118340/]
- 10. A High-Throughput Method for Specific, Rapid, Precise and ... [https://www.sciencedirect.com/science/article/abs/pii/S0039914025015681]
- 11. Enhanced Purification of Ubiquitinated Proteins by ... [https://www.sciencedirect.com/science/article/pii/S1535947620336288]
- 12. Tools for Decoding Ubiquitin Signaling in DNA Repair - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC8690248/]
- 13. Ubiquitin diGLY proteomics as an approach to identify and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6791129/]
- 14. Rapid and deep-scale ubiquitylation profiling for biology ... [https://www.nature.com/articles/s41467-019-14175-1]
- 15. Current methodologies in protein ubiquitination characterization [https://cellandbioscience.biomedcentral.com/articles/10.1186/s13578-022-00870-y]
- 16. Frontiers | PROTAC : a revolutionary technology propelling small... [https://www.frontiersin.org/journals/oncology/articles/10.3389/fonc.2025.1676414/full]
- 17. Proteolysis‐Targeting Chimera (PROTAC) - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC12495453/]
- 18. Degraders in Clinical Trails: PROTAC Update | Biopharma PEG 2025 [https://www.biochempeg.com/article/434.html]
- 19. The latest trends in drug discovery | CAS [https://www.cas.org/resources/cas-insights/2025-drug-discovery-trends]
- 20. (IP) and co- Immunoprecipitation ... | Abcam immunoprecipitation [https://www.abcam.com/en-us/technical-resources/protocols/immunoprecipitation]
- 21. - Anti | Invitrogen Ubiquitin Antibodies [https://www.thermofisher.com/antibody/primary/target/ubiquitin]
- 22. Alzheimer's lesions labelled by anti - ubiquitin : antibodies ... comparison [https://pubmed.ncbi.nlm.nih.gov/7694167/]
- 23. Pan-cancer landscape of UBD/FAT10 and experimental ... [https://www.frontiersin.org/articles/10.3389/fonc.2025.1615898/full]
- 24. Super Enhanced Purification of Denatured-Refolded ... [https://www.sciencedirect.com/science/article/pii/S1535947624001427]
- 25. Enrichment and Expansion of Cells using Antibody-Coated Micropallet Arrays - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC2753796/]
- 26. Microplate Selection and Recommended Practices in High ... [https://www.ncbi.nlm.nih.gov/books/NBK558077/]
- 27. Mass Spectrometry Sample Preparation Guide [https://www.organomation.com/mass-spectrometry-sample-preparation-guide]
- 28. Digital Microfluidics for Sample Preparation in Low‐Input ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11740955/]
- 29. Simple But Efficacious Enrichment of Integral Membrane ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9062332/]
- 30. Secondary interactions in ubiquitin-binding domains ... [https://www.sciencedirect.com/science/article/pii/S221112472400874X]
- 31. from ubiquitinated protein to ubiquitin chain architecture | Cell ... [https://cellandbioscience.biomedcentral.com/articles/10.1186/s13578-022-00870-y/tables/1]
- 32. Linker-specific monoclonal antibodies present a simple and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11403257/]
- 33. Ubiquiton—An inducible, linkage-specific ... [https://www.sciencedirect.com/science/article/pii/S1097276523009619]
- 34. Recent Trends and Innovations in Bead-Based Biosensors ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11086254/]
- 35. Bead-based approaches for increased sensitivity and ... [https://www.nature.com/articles/s41551-025-01498-2]
- 36. Affinity Enrichment for MS: Improving the yield of low ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6081742/]
- 37. Super Enhanced Purification of Denatured-Refolded ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11584597/]
- 38. IP vs. Affinity Chromatography vs. Pull-Down Assays [https://www.creative-proteomics.com/resource/comparative-analysis-protein-enrichment-techniques.htm]
- 39. Systematic assessment of antibody selectivity in plasma ... [https://www.nature.com/articles/s41598-019-43552-5]
- 40. Sequence enrichment profiles enable target-agnostic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10261905/]
- 41. Pan-cancer multi-omics profiling reveals ubiquitin D as a ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12480306/]
- 42. A versatile new tool derived from a bacterial deubiquitylase to ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9278747/]
- 43. Science Forum: Antibody characterization is critical to ... [https://elifesciences.org/articles/100211]
- 44. A Data Set of Human Endogenous Protein Ubiquitination ... [https://www.sciencedirect.com/science/article/pii/S1535947620302292]
- 45. Highly Accurate and Robust Absolute Quantification of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10581745/]
- 46. Comparison of Different Buffers for Protein Extraction from ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0142650]
- 47. Determining buffer conditions for downstream processing ... [https://www.nature.com/articles/s41598-023-37614-y]
- 48. Protein Adsorption on a Multimodal Cation Exchanger [https://www.mdpi.com/1420-3049/30/16/3389]
- 49. ELISA Coating Buffers | ImmunoChemistry Technologies [https://www.antibodiesinc.com/collections/coating-buffers?srsltid=AfmBOorLfh4NjvhrWpQC10A-6JUCMjhqJILRVHpbaDUryqvAXabuF15P]
- 50. - Non specific [https://www.sprpages.nl/best-results/artefacts/non-specific]
- 51. - Artifacts staining in flow cytometry, Part II - Sanguine and non specific [https://sanguinebio.com/artifacts-and-non-specific-staining-in-flow-cytometry-part-ii/]
- 52. The Origin and Implications of Artifact Ions in Bioanalytical ... [https://www.chromatographyonline.com/view/the-origin-and-implications-of-artifact-ions-in-bioanalytical-lc-ms]
- 53. Effective Sample Preparations in Imaging Mass Spectrometry [https://pmc.ncbi.nlm.nih.gov/articles/PMC4542606/]
- 54. Ubiquitin Chain Enrichment Middle-Down Mass ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5541364/]
- 55. What Is Used to Enrich Ubiquitin Proteins? [https://www.mtoz-biolabs.com/what-is-used-to-enrich-ubiquitin-proteins.html]
- 56. UBD Polyclonal Antibody (PA5-80201) [https://www.thermofisher.com/antibody/product/UBD-Antibody-Polyclonal/PA5-80201]
- 57. Ubiquitin-Like Protein UBD Promotes Cell Proliferation in ... [https://www.frontiersin.org/journals/oncology/articles/10.3389/fonc.2021.691347/full]
- 58. UBD‐mediated glycolytic reprogramming promotes M2 ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12278697/]
- 59. Structural basis of K11/K48-branched ubiquitin chain ... [https://www.nature.com/articles/s41467-025-64719-x]
- 60. Unraveling chain specific ubiquitination in cells using tandem ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12217038/]
- 61. Proteomic approaches to study ubiquitinomics [https://www.sciencedirect.com/science/article/abs/pii/S1874939923000354]
- 62. Ubiquitin D promotes the progression of rheumatoid ... [https://www.spandidos-publications.com/10.3892/mmr.2023.12940]
- 63. Antibody Purification Methods [https://www.thermofisher.com/us/en/home/life-science/antibodies/antibodies-learning-center/antibodies-resource-library/antibody-methods/antibody-purification-methods.html]
- 64. Technological advancements in antibody-based therapeutics ... [https://jbiomedsci.biomedcentral.com/articles/10.1186/s12929-025-01190-2]
- 65. A Cost–Benefit Analysis Simulation for the Digitalisation of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10141111/]
- 66. A novel approach for simulating and optimizing the ... [https://www.sciencedirect.com/science/article/pii/S2405844024169630]
- 67. Cost Benefit Analyses for Optimization Projects [https://princetonoptimization.com/blog/cost-benefit-analyses-optimization-projects/]