Ubiquiton: Mastering Linkage-Specific Polyubiquitylation to Decode Cellular Signals and Advance Therapeutics
The Ubiquiton system represents a transformative synthetic biology tool that enables rapid, inducible, and linkage-specific polyubiquitylation of proteins of interest in living cells.
Ubiquiton: Mastering Linkage-Specific Polyubiquitylation to Decode Cellular Signals and Advance Therapeutics
Abstract
The Ubiquiton system represents a transformative synthetic biology tool that enables rapid, inducible, and linkage-specific polyubiquitylation of proteins of interest in living cells. This review details its core mechanism—employing engineered E3 ligases and ubiquitin acceptor tags—and validates its application across diverse protein types, from soluble nuclear factors to integral membrane receptors. We explore how Ubiquiton empowers researchers to precisely dissect the functions of linear (M1-), K48-, and K63-linked ubiquitin chains in fundamental processes like proteasomal targeting and endocytosis. For an audience of researchers and drug development professionals, this article provides a comprehensive guide to implementing Ubiquiton, from foundational principles and methodological protocols to troubleshooting and benchmarking against existing techniques, thereby offering a robust framework to accelerate the study of ubiquitin signaling in health and disease.
Decoding the Ubiquitin Code: Why Linkage-Specific Tools Are a Game-Changer
The ubiquitin-proteasome system (UPS) represents a crucial regulatory mechanism for protein degradation and function in eukaryotic cells, governing nearly all cellular processes including cell cycle progression, signal transduction, and stress responses [1] [2]. This sophisticated system operates through a sequential enzymatic cascade that conjugates the small protein modifier ubiquitin to target substrates. The fundamental ubiquitination reaction involves three key enzyme classes: ubiquitin-activating (E1), ubiquitin-conjugating (E2), and ubiquitin-ligating (E3) enzymes [1]. The specificity and diversity of ubiquitin signaling emerge from the combinatorial interplay between these enzymes, particularly the extensive E3 ligase family which determines substrate selection. Deficiencies in this system are implicated in numerous human pathologies, including cancer, neurodegenerative disorders, and immune defects [3] [4]. This primer examines the core enzymatic machinery of the ubiquitin cascade while contextualizing its function within modern research applications, with particular emphasis on the innovative Ubiquiton toolset for linkage-specific polyubiquitylation.
The Enzymatic Cascade: Core Components and Mechanisms
E1 Activating Enzymes: Molecular Choreography of Ubiquitin Activation
The ubiquitination cascade initiates with E1 activating enzymes, which perform the ATP-dependent activation of ubiquitin through a two-step reaction [1]. First, E1 catalyzes the adenylation of ubiquitin's C-terminus, consuming ATP and producing pyrophosphate. Subsequently, the activated ubiquitin is transferred to the E1 catalytic cysteine residue, forming a high-energy thioester bond [1] [5]. This E1~Ub thioester intermediate represents the first energy-rich linkage in the cascade. Humans possess only two E1 enzymes for ubiquitin (UBE1 and UBA6), highlighting their broad specificity and foundational role [5]. Structural studies reveal that E1 enzymes undergo significant conformational changes during their catalytic cycle, transitioning between distinct states competent for ubiquitin adenylation, thioester formation, and eventual transfer to E2 conjugating enzymes [1]. This molecular choreography ensures the faithful initiation of ubiquitin signaling while maintaining strict specificity for different ubiquitin-like proteins (Ubls).
Table 1: Core Enzymes of the Ubiquitin Cascade
| Enzyme Class | Number in Humans | Core Function | Key Reaction | Representative Examples |
|---|---|---|---|---|
| E1 Activating | 2 | Ubiquitin activation via ATP hydrolysis | Ubiquitin adenylation, E1~Ub thioester formation | UBE1, UBA6 |
| E2 Conjugating | ~30 | Ubiquitin carriage and transfer | E2~Ub thioester formation, coordination with E3 | UBE2E1, UBE2L3, UBE2O |
| E3 Ligating | ~600 | Substrate recognition and specificity | Ubiquitin transfer to substrate lysine | RING-type, HECT-type, RBR-type |
E2 Conjugating Enzymes: Ubiquitin Carriers and Transfer Catalysts
E2 conjugating enzymes serve as the central carriers of activated ubiquitin, receiving it from E1 via trans-thioesterification and coordinating with E3 ligases for substrate modification [5]. The human genome encodes approximately 30 E2s, each containing a conserved ubiquitin-conjugating (UBC) catalytic domain that houses the active-site cysteine residue [6] [3]. E2 enzymes are categorized into four structural classes based on the presence of N-terminal and/or C-terminal extensions beyond the core UBC domain [6]. These extensions modulate subcellular localization, E1/E3 interactions, and functional specialization. Remarkably, certain E2s like UBE2O and BIRC6 function as E2/E3 hybrid enzymes that catalyze substrate ubiquitination independently of canonical E3 ligases [6]. Structural analyses of UBE2E1 have revealed its unique capacity for E3-independent ubiquitination when recognizing specific substrate sequences, demonstrating unexpected versatility within the E2 family [3].
E3 Ligases: Specificity Determinants in Ubiquitin Signaling
E3 ubiquitin ligases constitute the most diverse and specialized component of the cascade, with approximately 600 members in humans that determine substrate specificity and modification type [5] [3]. E3s are broadly classified into three major families based on their structural features and catalytic mechanisms: RING-type, HECT-type, and RBR-type E3 ligases. RING E3s function primarily as scaffolds that facilitate direct ubiquitin transfer from E2~Ub to substrates, while HECT and RBR E3s form catalytic thioester intermediates with ubiquitin before substrate modification [5]. The critical positioning function of E3s ensures precise lysine targeting on appropriate substrates, with their activity frequently regulated by post-translational modifications or allosteric effectors [2]. This extensive family enables the UPS to achieve remarkable substrate discrimination, allowing specific regulation of individual proteins within the complex cellular environment.
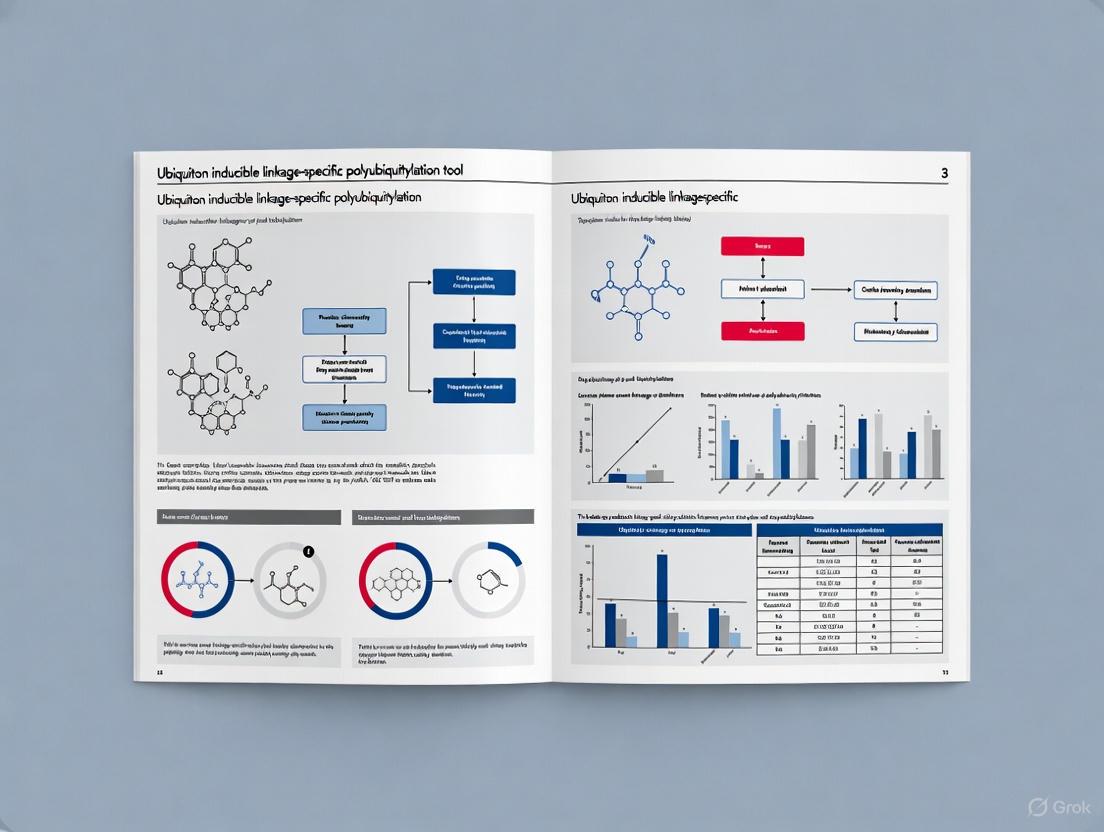
Ubiquiton: An Inducible, Linkage-Specific Polyubiquitylation Tool
The Ubiquiton system represents a groundbreaking synthetic biology approach for achieving precise, inducible control over protein polyubiquitylation in living cells [7] [8] [9]. This innovative toolset addresses a long-standing experimental limitation in ubiquitin research: the inability to induce specific ubiquitin chain linkages on proteins of interest within their native cellular environment. The system comprises engineered ubiquitin protein ligases paired with matching ubiquitin acceptor tags that together enable rapid, inducible formation of linear (M1-), K48-, or K63-linked polyubiquitin chains [7]. This design permits researchers to directly manipulate the ubiquitin code with unprecedented precision, facilitating mechanistic studies of chain-specific signaling outcomes without relying on the endogenous, often redundant, ubiquitination machinery.
Applications and Validation
The Ubiquiton system has been rigorously validated across diverse biological contexts and protein classes, demonstrating remarkable versatility. Researchers have successfully applied the tool to soluble cytoplasmic and nuclear proteins, chromatin-associated factors, and integral membrane proteins [7] [9]. Key applications include controlled proteasomal targeting of substrates via K48-linked chains and regulation of the endocytic pathway through K63-linked ubiquitination [7]. This flexibility enables investigation of ubiquitin signaling in virtually any cellular compartment or pathway. The inducible nature of the system allows precise temporal control, making it possible to trigger ubiquitination at specific timepoints and monitor subsequent phenotypic consequences, thus establishing clear cause-effect relationships that are often elusive in ubiquitin research.
Experimental Approaches and Methodologies
Quantitative Ubiquitinomics: Profiling the Ubiquitin Code
Modern ubiquitin research employs sophisticated quantitative proteomic approaches to comprehensively map ubiquitination events and quantify their dynamics [2] [4]. These methodologies typically combine enrichment strategies for ubiquitinated peptides with high-resolution mass spectrometry, enabling system-wide identification of ubiquitination sites and their relative abundances under different conditions. The standard workflow involves several critical steps: (1) protein extraction under denaturing conditions to preserve ubiquitination; (2) tryptic digestion to generate peptides; (3) immunoaffinity enrichment of ubiquitinated peptides using anti-diglycine (K-ε-GG) remnant antibodies; (4) LC-MS/MS analysis; and (5) bioinformatic processing and quantification [4]. For label-free quantification, signal intensity and spectral counting provide relative abundance measurements, while isobaric tagging approaches like TMT enable multiplexed analysis of multiple conditions [2]. These powerful methods have revealed startling complexity in ubiquitin signaling, with studies identifying up to 10,000 distinct ubiquitination sites in single experiments [4].
Activity-Based Profiling: Monitoring Enzymatic Cascades
Activity-based probes (ABPs) represent another powerful tool class for interrogating ubiquitin cascade dynamics, particularly for monitoring enzymatic activities rather than mere protein abundance [5]. The UbDha (UbGly76Dha) probe exemplifies this approach—a cascading ABP that mimics native ubiquitin but contains a C-terminal dehydroalanine moiety that covalently traps active-site cysteine residues of catalytically competent enzymes [5]. This innovative design enables the probe to "hop" through the entire E1-E2-E3 cascade while providing the option to irreversibly "trap" enzymes at each transfer step. The ATP-dependent reactivity of UbDha specifically reports on active enzymes rather than inactive pool components, making it ideal for profiling functional states of ubiquitin machinery in diverse physiological and pathological contexts [5].
Table 2: Key Research Reagent Solutions for Ubiquitin Cascade Studies
| Reagent / Tool | Type | Primary Function | Key Features & Applications |
|---|---|---|---|
| Ubiquiton System | Engineered ligases & tags | Inducible, linkage-specific polyubiquitylation | Enables M1-, K48-, K63-linked chains; works in yeast & mammalian cells |
| UbDha Probe | Activity-based probe | Monitoring active ubiquitin-conjugating enzymes | Traps E1, E2, HECT/RBR E3 active sites; reports on catalytic activity |
| Anti-K-ε-GG Antibody | Immunoaffinity reagent | Ubiquitinated peptide enrichment | Essential for ubiquitinomics; recognizes diglycine remnant on lysine |
| Linear Ubiquitin Antibody | Specific antibody | Detection of M1-linked chains | Clone LUB9; validates linear ubiquitination events |
| Linkage-Specific Antibodies | Specific antibodies | Detection of chain-type | e.g., Anti-Ubiquitin Lys63-Specific (Apu3 clone) |
E3-Independent Ubiquitination via UBE2E1 (SUE1 Method)
Recent structural and biochemical insights into the unique E2 enzyme UBE2E1 have enabled development of the SUE1 (sequence-dependent ubiquitination using UBE2E1) method for E3-free ubiquitination [3]. This approach exploits UBE2E1's natural ability to recognize specific hexapeptide sequences (originally identified in SETDB1 protein) and mediate direct ubiquitination without E3 involvement. The methodology involves several key steps: (1) identifying the minimal recognition sequence (KEGYES) required for UBE2E1 binding; (2) structural-guided optimization to enhance ubiquitination efficiency (KEGYEE); (3) introducing this optimized sequence into target proteins as a fusion tag; and (4) performing in vitro ubiquitination reactions with purified E1, UBE2E1, and ubiquitin [3]. This streamlined system efficiently generates ubiquitinated proteins with customized modification sites, ubiquitin chain linkages, and lengths, bypassing the challenge of identifying cognate E3 ligases for specific substrates.
Pathophysiological Connections and Therapeutic Implications
Dysregulation of the ubiquitin cascade contributes significantly to human disease, particularly in cancer where altered ubiquitination patterns drive oncogenic signaling, evade growth suppression, and resist cell death [4]. Quantitative ubiquitinomics of lung squamous cell carcinoma (LSCC) tissue has revealed profound alterations in ubiquitination patterns, with 627 differentially ubiquitinated proteins and 1209 modified lysine sites identified compared to adjacent normal tissue [4]. These modifications predominantly affect pathways controlling cell adhesion, signal transduction, ribosome function, and proteasome activity. Notably, KEGG pathway analysis identified 47 significantly altered signaling routes in LSCC, including mTOR, HIF-1, PI3K-Akt, and Ras pathways—all established cancer-associated networks [4]. These findings highlight the UPS as a rich source of potential diagnostic biomarkers and therapeutic targets, with several ubiquitin-pathway inhibitors already achieving clinical success (e.g., bortezomib, carfilzomib) for hematological malignancies [4].
The ubiquitin cascade, comprising E1, E2, and E3 enzymes, represents a sophisticated regulatory system of unparalleled importance in cellular homeostasis. While the fundamental mechanisms of ubiquitin activation, transfer, and ligation are now well-established, emerging tools like the Ubiquiton system are revolutionizing our ability to manipulate and study specific ubiquitination events with precision previously unimaginable. Combined with advanced proteomic methods and activity-based probes, these technologies are illuminating the intricate dynamics of ubiquitin signaling in health and disease. As research continues to unravel the complexity of this system, particularly through structural insights and innovative engineering approaches, our understanding of ubiquitin biology will undoubtedly expand, revealing new therapeutic opportunities for the numerous pathologies linked to UPS dysfunction.
Classical Ubiquitin Cascade Pathway - This diagram illustrates the sequential E1-E2-E3 enzymatic cascade that conjugates ubiquitin to protein substrates, with dashed lines indicating non-canonical E2/E3 hybrid enzyme pathways.
Ubiquiton Inducible Polyubiquitylation System - This diagram shows the engineered Ubiquiton system components and mechanism for achieving inducible, linkage-specific protein polyubiquitylation in living cells.
Ubiquitination is a fundamental post-translational modification that extends far beyond its initial characterization as a mere signal for proteasomal degradation. The covalent attachment of ubiquitin to substrate proteins creates a complex 'ubiquitin code'—a sophisticated language of biological regulation written in the form of polymeric chains of distinct architectures and linkage types [7] [10]. This code is deciphered by specialized effector proteins containing ubiquitin-binding domains, leading to diverse cellular outcomes including inflammatory signaling, DNA damage repair, transcriptional regulation, and endocytic trafficking [10] [11]. The functional diversity of ubiquitin signaling primarily stems from the ability of ubiquitin itself to form polymers through any of its seven lysine residues (K6, K11, K27, K29, K33, K48, K63) or its N-terminal methionine (M1), creating linkage-specific polyubiquitin chains that are recognized as distinct signals within the cell [10] [11]. Recent discoveries have further expanded this paradigm to include non-lysine ubiquitination events involving serine, threonine, and even phosphoribosyl linkages, as well as the critical regulatory functions of branched ubiquitin chains that combine multiple linkage types within a single polymeric structure [10] [12].
The Ubiquiton system represents a transformative methodological advancement for probing this complex ubiquitin code. This engineered tool enables rapid, inducible, and linkage-specific polyubiquitylation of target proteins in both yeast and mammalian cells, overcoming previous experimental limitations in studying defined ubiquitin signals in their native cellular environments [7]. By providing precise control over ubiquitin chain architecture on proteins of interest, Ubiquiton serves as an essential platform for deconvoluting the specific biological functions of different ubiquitin linkages, from the well-characterized K48-linked degradative signals to the non-proteolytic roles of K63-linked and linear ubiquitin chains.
Linkage-Specific Ubiquitin Functions: Application Notes
The functional spectrum of ubiquitin linkages encompasses both proteolytic and non-proteolytic signaling pathways, with distinct chain architectures triggering specific cellular responses. The following application notes detail the current understanding of key ubiquitin linkage types, with experimental data obtained through tools like the Ubiquiton system providing new mechanistic insights.
Table 1: Functional Diversity of Major Ubiquitin Linkages
| Linkage Type | Primary Functions | Key E3 Ligases | Cellular Processes | Experimental Tools |
|---|---|---|---|---|
| K48-linked | Proteasomal degradation [11] | UBR5 [12] | Protein turnover, cell cycle regulation | Ubiquiton-K48 system [7] |
| K63-linked | Non-proteolytic signaling [10] | Ubc13-Mms2 complex [10] | DNA repair, NF-κB activation, endocytosis | Ubiquiton-K63 system [7] |
| Linear (M1-linked) | Innate immune signaling [10] | LUBAC (HOIP/HOIL-1/SHARPIN) [10] | NF-κB activation, cell death regulation | Ubiquiton-M1 system [7] |
| K29-linked | Proteasomal degradation (in branched chains) [12] | TRIP12 [12] | ER-associated degradation, transcriptional regulation | TRABID-NZF1 binder [12] |
| K11-linked | Proteasomal degradation [12] | APC/C, UBR5 [12] | Cell cycle progression, ERAD | Linkage-specific antibodies [11] |
| K27/K33-linked | Less characterized immune signaling [11] | - | Immune response, mitophagy | Limited tools available |
| Branched (K29/K48) | Enhanced proteasomal targeting [12] | TRIP12 & UBR5 cooperation [12] | Overcoming DUB protection, rapid degradation | TUBE-based proteomics [12] |
K48-Linked Ubiquitin Chains: Beyond Simple Degradation Signals
K48-linked polyubiquitin chains represent the most abundant ubiquitin linkage type in mammalian cells and serve as the canonical signal for proteasomal degradation [11]. However, recent research using inducible ubiquitination tools has revealed unexpected subtleties in K48-chain function. When deployed using the Ubiquiton system against substrates of the endocytic pathway, K48-linked ubiquitylation not only triggered proteasomal degradation of soluble cytoplasmic proteins but also effectively targeted integral membrane proteins for internalization and destruction, demonstrating the versatility of this signal across cellular compartments [7]. The degradation efficiency of K48-linked chains can be significantly enhanced when incorporated into branched ubiquitin structures, particularly K29/K48 branched chains, which function as priority degradation signals that resist deubiquitylation and enhance proteasome recruitment [12].
The cooperative activity of E3 ligases UBR5 (K48-specific) and TRIP12 (K29-specific) in generating K29/K48 branched chains on substrates like OTUD5 represents a sophisticated mechanism for ensuring robust degradation of otherwise protected proteins [12]. This branched architecture overcomes the deubiquitylating activity of OTUD5, which readily cleaves K48 linkages but has limited efficacy against K29 linkages, creating a DUB-resistant degradation signal that shifts the ubiquitin conjugation/deconjugation equilibrium toward substrate destruction [12].
K63-Linked and Linear Ubiquitin Chains: Non-Proteolytic Signaling Hubs
K63-linked ubiquitin chains serve as critical non-degradative signaling scaffolds in multiple cellular pathways, with structural studies revealing how the Ubc13-Mms2 heterodimer specifically orients the acceptor ubiquitin to facilitate K63 linkage formation [10]. Application of the Ubiquiton-K63 system to chromatin-associated proteins has demonstrated how this linkage type regulates transcriptional activation and DNA damage repair through mechanisms involving recruitment of specific repair complexes and modification of histone proteins [7] [10]. Similarly, linear (M1-linked) ubiquitin chains assembled by the LUBAC complex (HOIP/HOIL-1/SHARPIN) function as essential scaffolds in innate immune signaling, creating recruitment platforms for downstream effectors in the NF-κB pathway [10]. The Ubiquiton system for linear chains provides a valuable tool for dissecting the specific contributions of linear ubiquitination without simultaneously activating other LUBAC-dependent signaling events.
Branched Ubiquitin Chains: Enhanced Degradation Signals
Branched ubiquitin chains containing multiple linkage types represent a recently appreciated layer of complexity in the ubiquitin code that function as priority signals for proteasomal degradation [12]. The K29/K48 branched ubiquitin chains assembled by the cooperative action of TRIP12 and UBR5 on OTUD5 create a superior degradation signal that enhances proteasome recruitment and resists deubiquitylation [12]. This mechanism is particularly important for the degradation of DUB-protected substrates, as the DUB-resistant K29 linkages provide a foundation for K48-linked branching that ultimately overwhelms the substrate's protective deubiquitylation capacity. Similar principles apply to K11/K48 and K48/K63 branched chains, which also demonstrate enhanced degradation efficiency compared to their homotypic counterparts [12].
Experimental Protocols: Probing Linkage-Specific Functions
Protocol: Inducible Linkage-Specific Ubiquitylation Using Ubiquiton System
Purpose: To induce rapid, specific polyubiquitin chain formation on a protein of interest to study linkage-specific functions [7].
Materials:
- Ubiquiton plasmid set (engineered E3 ligases and matching ubiquitin acceptor tags)
- Target gene construct with appropriate tagging
- Cell line of interest (yeast or mammalian)
- Inducer (e.g., doxycycline or other small-molecule inducer)
- Proteasome inhibitor (e.g., MG132)
- Linkage-specific ubiquitin antibodies (e.g., anti-K63 [Apu3], anti-linear [LUB9]) [9]
- General ubiquitin detection antibodies (e.g., FK2) [9]
Methodology:
- Molecular Cloning: Fuse the target protein gene with the appropriate Ubiquiton acceptor tag using standard molecular biology techniques.
- Cell Line Generation: Co-transfect cells with the tagged target construct and the appropriate engineered E3 ligase plasmid specific for the desired linkage (K48, K63, or M1).
- Induction of Ubiquitylation: Treat cells with the specific inducer (concentration and time must be optimized for the experimental system) to initiate polyubiquitin chain formation.
- Validation of Ubiquitylation:
- Harvest cells at appropriate time points post-induction
- Prepare cell lysates with protease inhibitors (e.g., PMSF, protease inhibitor cocktail) [9]
- Perform immunoblotting with linkage-specific antibodies to confirm formation of the desired ubiquitin chain type
- Functional Assays: Assess downstream consequences using appropriate assays:
- Protein stability assays (cycloheximide chase) [9]
- Subcellular localization (immunofluorescence)
- Interaction studies (co-immunoprecipitation)
- Pathway-specific readouts (e.g., NF-κB activation for linear chains)
Troubleshooting Notes:
- Incomplete ubiquitylation may require optimization of inducer concentration or timing
- Verify linkage specificity using linkage-specific antibodies and ubiquitin mutants
- Include controls with catalytically inactive E3 ligase variants
Protocol: Identification of Branched Ubiquitin Chain Substrates
Purpose: To identify and validate cellular substrates modified with K29/K48 branched ubiquitin chains using proteomic and biochemical approaches [12].
Materials:
- TRIP12 and UBR5 expression constructs
- siRNA for TRIP12/UBR5 knockdown
- Tandem ubiquitin-binding entities (TUBE2) for ubiquitin enrichment [12]
- GST-fused TRABID-NZF1 as K29 linkage binder [12]
- K48-linkage specific antibody [11]
- Mass spectrometry reagents and instrumentation
Methodology:
- Candidate Identification:
- Perform TRIP12 knockdown in target cells (e.g., HT1080 cells)
- Conduct TMT-based quantitative proteomics to identify accumulated proteins
- Validate candidates by immunoblotting after TRIP12 depletion
- Ubiquitin Linkage Characterization:
- Enrich ubiquitinated proteins using TUBE2 affinity purification
- Use linkage-specific binders (TRABID-NZF1 for K29) to isolate specific chain types
- Confirm K29/K48 branched chains using Ub-AQUA/PRM mass spectrometry
- Functional Validation:
- Measure protein half-life after TRIP12/UBR5 perturbation
- Assess ubiquitylation in vitro with purified components
- Determine functional consequences in relevant pathways (e.g., NF-κB signaling for OTUD5)
Applications: This protocol has been successfully applied to identify OTUD5 as a substrate for TRIP12/UBR5-mediated K29/K48 branched ubiquitylation, revealing a mechanism for overcoming DUB-protected degradation [12].
Research Reagent Solutions: Essential Tools for Ubiquitin Research
Table 2: Key Research Reagents for Studying Ubiquitin Linkages
| Reagent / Tool | Specificity / Function | Key Applications | Examples / Sources |
|---|---|---|---|
| Ubiquiton System | Inducible, linkage-specific polyubiquitylation | Controlled study of specific ubiquitin chain functions in cells | Engineered E3 ligases for K48, K63, M1 linkages [7] |
| Linkage-Specific Antibodies | Recognize specific ubiquitin chain types | Detection and validation of chain linkage in immunoblotting | Apu3 (K63-specific), LUB9 (linear), K48-specific antibodies [11] [9] |
| Tandem UBD Probes | High-affinity ubiquitin binders (pan-specific or linkage-selective) | Enrichment of ubiquitinated proteins from complex mixtures | TUBE2 (pan-specific), TRABID-NZF1 (K29/K33-selective) [12] |
| Ubiquitin Variants | Mutant ubiquitin with specific lysine residues mutated | Determining linkage specificity in ubiquitylation assays | Single-lysine ubiquitin mutants (e.g., K48R, K63R) [12] |
| Activity-Based Probes | Chemical tools for profiling DUB activity and specificity | Characterizing deubiquitylase functions and specificity | Transthiolation activity profiling probes [10] |
| Proteasome Inhibitors | Block proteasomal degradation | Stabilizing ubiquitinated proteins for detection | MG132, bortezomib, carfilzomib |
| E3 Ligase Modulators | Activate or inhibit specific E3 ligases | Functional perturbation of ubiquitination pathways | TRIP12/UBR5 modulators for branched chain studies [12] |
Signaling Pathway Visualizations
K29/K48 Branched Ubiquitin Pathway for DUB-Protected Degradation
Ubiquiton Experimental Workflow for Inducible Ubiquitylation
The Ubiquiton inducible linkage-specific polyubiquitylation system represents a transformative methodological advancement that enables precise dissection of the ubiquitin code's functional complexity. By moving beyond generic ubiquitination studies to linkage-specific manipulation, this tool provides unprecedented insight into how different ubiquitin chain architectures dictate diverse cellular outcomes. The emerging understanding of branched ubiquitin chains as priority degradation signals that overcome DUB protection highlights the sophistication of this regulatory system and opens new therapeutic avenues for targeting previously undruggable proteins. As research continues to unravel the complexities of the ubiquitin code, including non-canonical ubiquitination events and the crosstalk between different linkage types, tools like Ubiquiton will be essential for establishing causal relationships between specific ubiquitin signals and their biological consequences, ultimately advancing both basic science and drug discovery in the ubiquitin field.
The ubiquitin-proteasome system (UPS) represents a crucial post-translational modification pathway that governs diverse cellular functions, with its specificity largely determined by the topology of polyubiquitin chains. Among the eight distinct linkage types, K48-linked chains primarily target substrates for proteasomal degradation, while K63-linked chains predominantly regulate non-proteolytic processes including signal transduction, protein trafficking, and inflammatory pathway activation [13] [14]. Despite this understanding, the functional characterization of specific ubiquitin linkages has been severely hampered by technological limitations in traditional experimental approaches. This application note delineates the critical methodological constraints inherent in conventional ubiquitination studies and presents advanced tools and protocols that bridge this experimental gap, with particular emphasis on their application within Ubiquiton-inducible linkage-specific polyubiquitylation research.
The Limitations of Conventional Methodologies
Traditional approaches for studying linkage-specific ubiquitination suffer from several significant shortcomings that affect their sensitivity, throughput, and biological relevance.
Key Methodological Constraints
- Mass Spectrometry: While powerful for identifying ubiquitination sites, this method is labor-intensive, requires sophisticated instrumentation, and lacks the sensitivity to capture rapid, transient changes in endogenous protein ubiquitination within cellular contexts [13].
- Mutant Ubiquitin Expression: Techniques relying on exogenous expression of ubiquitin mutants (where lysines are mutated to arginine) may not accurately recapitulate modifications involving wild-type ubiquitin, potentially introducing artifacts and compromising physiological relevance [13].
- Western Blotting: The standard workhorse for protein detection is low-throughput, provides only semi-quantitative data, and often lacks the sensitivity required to detect subtle but biologically significant changes in ubiquitination states [13].
These limitations collectively create a significant experimental gap, impeding rapid progress in the UPS field, particularly in drug discovery endeavors such as the characterization of Proteolysis Targeting Chimeras (PROTACs) and molecular glues [13].
Table 1: Quantitative Comparison of Traditional vs. Advanced Ubiquitination Assessment Methods
| Methodological Parameter | Western Blotting | Mass Spectrometry | Mutant Ubiquitin | TUBEs-Based Assay | Ubiquiton System |
|---|---|---|---|---|---|
| Throughput | Low | Low | Medium | High (96/384-well) | High |
| Quantitative Capability | Semi-quantitative | Quantitative | Semi-quantitative | Quantitative | Tunable/Kinetic |
| Linkage Specificity | Limited (via Ab) | High | High (by design) | High (K48, K63, Pan) | High (M1, K48, K63) |
| Endogenous Context | Yes | Yes | No (overexpression) | Yes | Inducible |
| Sensitivity | Low to Moderate | Moderate | Variable | High (nanomolar affinity) | High |
| Experimental Workflow Complexity | Moderate | High | Moderate | Low to Moderate | Moderate (setup) |
Advanced Tools to Bridge the Gap
The development of sophisticated molecular tools has begun to address the limitations of traditional methods, enabling precise, linkage-specific investigation of ubiquitination.
Chain-Specific Tandem Ubiquitin Binding Entities (TUBEs)
TUBEs are engineered affinity matrices with nanomolar affinities for specific polyubiquitin chains. Their application in high-throughput screening (HTS) assays allows for the precise capture and analysis of linkage-specific ubiquitination events on native proteins [13]. For instance, K63-TUBEs selectively capture L18-MDP-induced inflammatory signaling ubiquitination of RIPK2, while K48-TUBEs specifically bind RIPK2 PROTAC-induced degradative ubiquitination, enabling clear functional differentiation [13].
The Ubiquiton Inducible Polyubiquitylation System
The Ubiquiton system represents a groundbreaking synthetic biology tool for controlling ubiquitination. It comprises a set of engineered E3 ubiquitin ligases and matching ubiquitin acceptor tags that enable rapid, inducible, and linkage-specific (M1, K48, or K63) polyubiquitylation of proteins of interest in both yeast and mammalian cells [15] [7]. This system has been validated for controlling the localization and stability of diverse targets, including soluble cytoplasmic, nuclear, chromatin-associated, and integral membrane proteins [7].
Detailed Experimental Protocols
Protocol 1: Assessing Linkage-Specific Endogenous Ubiquitination Using TUBEs
This protocol outlines the procedure for capturing and detecting linkage-specific ubiquitination of endogenous proteins, such as RIPK2, from cell lysates, utilizing the example from the search results [13].
- Application: Investigation of endogenous target protein ubiquitination (e.g., stimulus-induced K63 vs. PROTAC-induced K48 ubiquitination).
- Principle: Chain-specific TUBEs immobilized on magnetic beads selectively enrich for proteins modified with specific ubiquitin linkages from complex cell lysates.
Procedure:
- Cell Stimulation and Lysis:
- Culture THP-1 human monocytic cells under standard conditions.
- Pre-treat cells with compound of interest (e.g., 100 nM Ponatinib for RIPK2 inhibition) or vehicle control (DMSO) for 30 minutes [13].
- Stimulate cells with relevant agent (e.g., 200-500 ng/mL L18-MDP for K63 ubiquitination of RIPK2) or RIPK2 PROTAC (e.g., RIPK degrader-2 for K48 ubiquitination) for desired time (e.g., 30 min) [13].
- Lyse cells using a specialized lysis buffer (e.g., containing 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 10% Glycerol, and supplemented with 1x protease inhibitors and 10 mM N-Ethylmaleimide to preserve polyubiquitination) [13].
- TUBEs Affinity Enrichment:
- Clarify cell lysates by centrifugation at 15,000 x g for 15 minutes at 4°C.
- Quantify protein concentration. Use 500 µg - 1 mg of total protein lysate per enrichment condition.
- Incubate the clarified lysate with chain-specific TUBE-conjugated magnetic beads (e.g., K48-TUBE, K63-TUBE, or Pan-TUBE) for 2 hours at 4°C with gentle rotation [13].
- Washing and Elution:
- Pellet beads using a magnetic rack and carefully remove the supernatant.
- Wash beads three times with 1 mL of ice-cold lysis buffer without detergents.
- Elute bound proteins by boiling the beads in 1x Laemmli sample buffer for 10 minutes.
- Detection and Analysis:
- Resolve eluted proteins by SDS-PAGE and transfer to a PVDF membrane.
- Perform immunoblotting using a target-specific antibody (e.g., anti-RIPK2) to detect the ubiquitinated forms, which will appear as high-molecular-weight smears [13].
Protocol 2: Inducing Linkage-Specific Ubiquitination with the Ubiquiton System
This protocol describes the implementation of the Ubiquiton system to induce defined ubiquitin linkages on a protein of interest, facilitating the study of chain-specific outcomes.
- Application: Controlled, inducible polyubiquitylation of a target protein to study the consequences of specific linkages (e.g., proteasomal degradation for K48, endocytic trafficking for K63).
- Principle: A protein of interest is tagged with the Ubiquiton acceptor tag. Co-expression with an engineered E3 ligase specific for a desired linkage (M1, K48, K63) allows for the inducible recruitment and assembly of defined ubiquitin chains upon ligand addition.
Procedure:
- System Construction:
- Subclone the cDNA of your protein of interest (POI) into the appropriate Ubiquiton vector to create an N- or C-terminal fusion with the Ubiquiton acceptor tag.
- Choose the matching plasmid encoding the engineered E3 ligase for your desired linkage (M1-, K48-, or K63-specific).
- Cell Transfection and Induction:
- Co-transfect the POI-Ubiquiton tag plasmid and the selected engineered E3 ligase plasmid into your mammalian cell line of choice (e.g., HEK293T).
- Culture transfected cells for 24-48 hours to allow for protein expression.
- Induce polyubiquitylation by adding the specific small-molecule inducer (ligand) as per the Ubiquiton system specifications. The induction time will vary based on the experimental goal (e.g., short induction for degradation kinetics).
- Downstream Analysis:
- Analyze cells by Western blot to assess POI stability/degradation (K48) or modification.
- Employ immunofluorescence to monitor changes in subcellular localization (e.g., membrane recruitment for K63).
- Perform functional assays relevant to the induced ubiquitination (e.g., NF-κB reporter assays for K63/M1-linked signaling, or cell viability assays for essential protein degradation).
Visualizing Signaling and Experimental Workflows
Diagram 1: K63 vs K48 Ubiquitin Signaling Pathways in Inflammation.
Diagram 2: TUBEs-Based Enrichment Workflow for Endogenous Ubiquitination.
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagent Solutions for Linkage-Specific Ubiquitination Research
| Reagent / Tool | Specific Example | Function & Application |
|---|---|---|
| Chain-Specific TUBEs | K48-TUBE, K63-TUBE, Pan-TUBE (LifeSensors) | High-affinity capture and enrichment of proteins modified with specific ubiquitin linkages from native cell lysates for downstream detection [13]. |
| Inducible Ubiquitination System | Ubiquiton System (Engineered E3 ligases & acceptor tags) | Enables precise, rapid, and inducible formation of M1-, K48-, or K63-linked polyubiquitin chains on a protein of interest to study causal effects [15] [7]. |
| Ubiquitination-Preserving Lysis Buffer | 50 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 10% Glycerol, 10 mM N-Ethylmaleimide | Maintains polyubiquitin chains on substrate proteins during cell lysis by inhibiting deubiquitinases (DUBs) [13]. |
| Prototypical Inducers | L18-MDP (for NOD2/RIPK2 K63 ubiquitination), RIPK2 PROTAC (e.g., Degrader-2 for K48 ubiquitination) | Tool compounds used to stimulate specific endogenous ubiquitination pathways for mechanistic studies [13]. |
| Specific Inhibitors | Ponatinib (RIPK2 inhibitor), DUB inhibitors (e.g., specific for OTULIN, CYLD, A20) | Used to probe the requirement of specific enzymes in ubiquitination pathways and validate findings [13] [14]. |
| Copper L-aspartate | ||
| Musk Xylene-d9 | Musk Xylene-d9, MF:C12H15N3O6, MW:306.32 g/mol | Chemical Reagent |
Ubiquitination is a fundamental post-translational modification that regulates virtually every cellular process, controlling protein stability, activity, and localization [16]. The functional diversity of ubiquitination stems from the ability of ubiquitin to form polymeric chains through different internal lysine residues, creating distinct linkage types that encode specific cellular signals [7]. For decades, researchers have struggled to decipher this "ubiquitin code" due to a critical technological gap: the lack of tools to induce specific polyubiquitin linkages on proteins of interest in living cells [7] [8]. Conventional methods using enzymatic cascades or in vitro approaches cannot achieve precise linkage-specific control in cellular environments, significantly limiting our understanding of ubiquitin signaling.
The Ubiquiton system represents a transformative solution to this longstanding challenge. This innovative synthetic biology tool enables researchers for the first time to induce linkage-specific polyubiquitylation of target proteins with temporal control in both yeast and mammalian cells [7] [15]. By providing precision manipulation of ubiquitin signaling, Ubiquiton opens new avenues for investigating the roles of specific ubiquitin linkages in proteostasis, cellular signaling, and disease mechanisms, potentially accelerating drug discovery in areas ranging from cancer to neurodegenerative disorders.
Ubiquiton System Architecture: Engineered Components for Precision Targeting
Core System Components
The Ubiquiton system employs a modular design consisting of engineered ubiquitin protein ligases and matching ubiquitin acceptor tags that work in concert to achieve specific polyubiquitin chain formation.
Table: Core Components of the Ubiquiton System
| Component Type | Function | Specificity Options |
|---|---|---|
| Engineered E3 Ubiquitin Ligases | Catalyze attachment of ubiquitin to target proteins | Linear (M1-), K48-, or K63-linked polyubiquitylation |
| Ubiquitin Acceptor Tags | Genetically fused to proteins of interest for targeted modification | Compatible with various linkage-specific E3 ligases |
| Induction Mechanism | Enables temporal control of polyubiquitin chain formation | Rapid induction upon system activation |
The system has been rigorously validated for diverse protein types including soluble cytoplasmic and nuclear proteins, chromatin-associated factors, and integral membrane proteins [7] [15]. This broad applicability makes Ubiquiton particularly valuable for studying ubiquitination in various cellular contexts.
Molecular Mechanism of Action
The Ubiquiton system operates through a coordinated mechanism where engineered E3 ligases recognize and modify specific acceptor tags on target proteins. Upon induction, these customized ligases catalyze the formation of defined ubiquitin chain architectures, enabling researchers to precisely control the ubiquitination state of proteins of interest and observe subsequent cellular effects [8]. The system's design allows for inducible control, meaning ubiquitination can be initiated at specific time points to study dynamic cellular processes [7].
Quantitative System Validation: Proteomic Evidence for Specificity
Linkage Specificity and Proteomic Profiling
Rigorous quantitative proteomics has confirmed Ubiquiton's ability to generate specific polyubiquitin linkages with minimal off-target effects. SILAC-based and label-free quantitative proteomics experiments were conducted to verify the linkage specificity of polyubiquitin chains produced on model substrates like GFP in both yeast and human cells [17].
Table: Quantitative Proteomic Validation of Ubiquiton Specificity
| Validation Metric | Methodology | Key Finding |
|---|---|---|
| Linkage Specificity | SILAC-based proteomics | Confirmed precise formation of intended linkages (M1, K48, K63) |
| Off-target Assessment | Label-free quantitative proteomics | Minimal non-specific ubiquitination events detected |
| Cross-species Functionality | Comparative analysis in yeast and human cells | System functionality conserved across species |
| Substrate Scope | Multiple model substrates (e.g., GFP) | Consistent linkage specificity across different target proteins |
These proteomic analyses demonstrated that Ubiquiton achieves unprecedented specificity in generating defined ubiquitin chain types while maintaining minimal off-target effects, addressing a critical limitation of previous approaches to studying ubiquitination [17].
Application Notes: Controlled Protein Degradation and Membrane Trafficking
Precision Control of Protein Stability
The Ubiquiton system enables precise manipulation of protein half-lives through inducible K48-linked polyubiquitination, which primarily targets proteins for proteasomal degradation. This application is particularly valuable for:
- Studying Essential Genes: Conditionally deplete essential proteins to investigate their functions
- Signal Transduction Analysis: Control the stability of signaling components to map pathway dynamics
- Drug Target Validation: Mimic pharmacological inhibition by rapidly removing target proteins
Experimental validation has confirmed that Ubiquiton-induced K48-linked ubiquitination effectively directs substrates to the proteasome for degradation, providing researchers with a powerful tool for controlling protein abundance [7].
Regulation of Membrane Protein Trafficking
Beyond proteasomal targeting, Ubiquiton has been successfully applied to control endocytic trafficking of membrane proteins, including the Epidermal Growth Factor Receptor (EGFR) [7] [15]. This application leverages specific ubiquitin linkages known to regulate membrane protein internalization and sorting, allowing precise manipulation of:
- Receptor Endocytosis: Induce specific ubiquitin chains to control receptor internalization rates
- Intracellular Trafficking: Direct proteins to specific cellular compartments through linkage-specific ubiquitination
- Signal Modulation: Regulate signaling duration and intensity by controlling membrane protein localization
The successful application to EGFR demonstrates Ubiquiton's capability to modify even complex integral membrane proteins, significantly expanding the tool's utility beyond soluble substrates [15].
Experimental Protocols: Implementation Guidelines
System Implementation Workflow
The following diagram illustrates the key steps for implementing the Ubiquiton system in mammalian cells:
Ubiquitination Detection and Validation
Following system implementation, rigorous detection and validation are essential. The protocol below adapts established ubiquitination detection methods for use with the Ubiquiton system [18]:
Sample Preparation:
- Harvest cells at appropriate time points post-induction
- Lyse cells in buffer containing protease inhibitors and deubiquitinase (DUB) inhibitors (e.g., 1 mM iodoacetamide) to preserve ubiquitination
- Process samples for downstream analysis
Ubiquitination Detection:
- Separate proteins by SDS-PAGE under denaturing conditions
- Transfer to membranes and probe with:
- Anti-ubiquitin antibodies (to detect total ubiquitination)
- Linkage-specific ubiquitin antibodies (to verify chain type)
- Antibodies against your protein of interest (to detect ubiquitinated forms)
Functional Validation:
- For degradation studies: Assess protein half-life using cycloheximide chase assays
- For localization studies: Employ immunofluorescence or live-cell imaging
- For functional assays: Perform pathway-specific readouts relevant to your biological question
This protocol can be modified based on specific experimental needs, but should always include appropriate controls to verify linkage specificity and rule of off-target effects.
Research Reagent Solutions: Essential Tools for Implementation
Table: Key Research Reagents for Ubiquiton Applications
| Reagent Category | Specific Examples | Applications and Functions |
|---|---|---|
| Ubiquitin System Enzymes | E1 activating enzyme, E2 conjugating enzymes, Engineered E3 ligases | Essential components for ubiquitination cascade; Ubiquiton provides specialized E3 ligases [18] |
| Detection Antibodies | Anti-ubiquitin (P4D1, FK1/FK2), Linkage-specific ubiquitin antibodies, Anti-substrate antibodies | Verify ubiquitination, determine chain linkage type, detect target protein [16] |
| Affinity Purification Reagents | GST-qUBA tandem domains, Ni-NTA resin (for His-tagged Ub), Strep-Tactin (for Strep-tagged Ub) | Enrich ubiquitinated proteins for proteomic analysis or validation [19] [16] |
| Inhibitors | MG132 (proteasome inhibitor), Iodoacetamide (DUB inhibitor), 1,10-o-phenanthroline (DUB inhibitor) | Stabilize ubiquitinated proteins by blocking degradation and deubiquitination [20] [19] |
| Mass Spectrometry Supplies | LTQ-Orbitrap instruments, C18 columns, Trypsin for digestion | Identify ubiquitination sites via detection of diGly remnant (114.043 Da mass shift) [19] [20] |
Concluding Perspectives: Future Directions and Applications
The Ubiquiton system represents a paradigm shift in ubiquitin research, providing unprecedented precision in manipulating ubiquitin signaling. While the current toolset covers the most extensively studied linkages (M1, K48, K63), future developments will likely expand to encompass atypical ubiquitin linkages such as K6, K11, K27, K29, and K33, whose functions are less characterized but increasingly recognized as biologically important [20] [16].
The integration of Ubiquiton with emerging technologies in chemical biology [21] and advanced proteomics [20] [16] promises to further accelerate our decoding of the ubiquitin code. As this tool sees broader adoption, it will undoubtedly generate fundamental insights into cellular regulation and open new therapeutic avenues for human diseases characterized by ubiquitin pathway dysregulation, ultimately fulfilling the promise of precision control in biological research and therapeutic development.
A Practical Guide to Implementing the Ubiquiton System in Your Research
Within the burgeoning field of targeted protein ubiquitination, the inducible "Ubiquiton" system represents a significant synthetic biology advance. This platform enables precise, linkage-specific polyubiquitylation to direct fundamental cellular processes [15]. The core technological elements empowering this tool are engineered E3 ligases and ubiquitin acceptor tags. These components work in concert to overcome the inherent complexity of the native ubiquitination cascade, providing researchers with unprecedented control over protein fate. This document details the key reagents, quantitative performance metrics, and standardized protocols for implementing these systems, providing a essential resource for therapeutic development.
Engineered E3 Ligase Platforms
E3 ligases are the central specificity determinants in the ubiquitin-proteasome system. Recent engineering efforts have produced several versatile platforms that facilitate targeted ubiquitination for both basic research and drug discovery.
The BioE3 Substrate Identification Platform
The BioE3 platform is a powerful strategy designed to identify specific substrates for a given E3 ligase, a traditionally challenging task. It couples a biotin ligase (BirA)-E3 fusion protein with a bioGEF-tagged, non-cleavable ubiquitin (bioGEFUb(^nc)) to enable proximity-dependent biotinylation of ubiquitinated substrates, allowing for their streptavidin-based purification and identification via liquid chromatography-mass spectrometry (LC-MS) [22].
- Key Innovation: The use of a low-affinity AviTag variant (bioGEF) is critical. It replaces the canonical bioWHE sequence to minimize non-specific, background biotinylation, ensuring that labeling occurs only when the bioGEFUb is in close proximity to the BirA-E3 fusion during the ubiquitination event [22].
- Experimental Workflow: The core protocol involves generating a stable cell line (e.g., HEK293FT, U2OS) with a doxycycline (DOX)-inducible bioGEFUb(^nc) construct. These cells are cultured in biotin-depleted media before transfection with a BirA-E3 fusion construct. Simultaneous DOX induction and the addition of exogenous biotin then enable a time-limited labeling window, facilitating the capture of bona fide E3 substrates [22].
Diagram 1: BioE3 workflow for identifying E3 ligase substrates.
Engineered SCF (eSCR) E3 Ligases
Multisubunit SCF (SKP1/Cullin1/F-box) E3 ligases are challenging to reconstitute in vitro. A breakthrough solution involves engineering a fused SKP1-Cullin1-RBX1 protein (eSCR) combined with interchangeable F-box proteins that confer substrate specificity [23].
- Key Innovation: The eSCR fusion protein simplifies the production of an active E3 ligase core. By co-expressing this core with various F-box proteins (e.g., rice D3, GID2, or human FBXL18, CDC4), researchers can rapidly reconstitute a diverse array of functional SCF E3 ligases for in vitro ubiquitination assays [23].
- Application: This platform has been successfully used to ubiquitinate key signaling proteins, such as the D53 transcriptional repressor in strigolactone signaling, providing a powerful and modular system for studying the mechanisms of multisubunit E3 ligases [23].
HECT-family E3s for Branched Ubiquitination
Structural biology has illuminated the mechanism of HECT-type E3 ligases in assembling complex ubiquitin codes. Cryo-EM structures of the HECT-E3 Ufd4, and its human homolog TRIP12, reveal how it preferentially synthesizes K29-linked ubiquitin chains onto pre-existing K48-linked chains, forming K29/K48-branched ubiquitin chains [24].
- Key Insight: The N-terminal ARM region and HECT domain C-lobe of Ufd4 work together to recruit K48-linked diUb and orient Lys29 of the proximal Ub for catalysis. This branched chain topology acts as an enhanced degradation signal [24].
- Quantitative Specificity: Enzyme kinetics revealed Ufd4 has a ~5.2-fold higher catalytic efficiency ((k{cat}/Km)) for the proximal K29 site (0.11 µMâ»Â¹ minâ»Â¹) compared to the distal K29 site (0.021 µMâ»Â¹ minâ»Â¹) within a K48-linked diUb substrate [24].
Table 1: Catalytic Efficiency of Ufd4 on K48-Linked DiUb Substrates
| Ubiquitination Site on K48-diUb | kcat/Km (µMâ»Â¹ minâ»Â¹) | Relative Efficiency |
|---|---|---|
| Proximal Ub K29 | 0.11 | 5.2x |
| Distal Ub K29 | 0.021 | 1x (reference) |
Source: Adapted from [24]
Sequence-Dependent Ubiquitin Acceptor Tags
A paradigm-shifting approach bypasses the need for E3 ligases entirely by engineering substrate sequences that are directly recognized by specific E2 enzymes.
SUE1: E3-Free Ubiquitination using UBE2E1
The SUE1 (Sequence-dependent Ubiquitination using UBE2E1) platform leverages the unique ability of the human E2 enzyme UBE2E1 to catalyze site-specific monoubiquitination of a substrate hexapeptide (KEGYES) independently of an E3 ligase [3].
- Structural Mechanism: The crystal structure of UBE2E1 bound to the substrate peptide revealed an "L"-shaped conformation. Key residues (Y4 and E5) in the peptide act as anchor points, binding UBE2E1 residues P164 and S126, respectively, to position the target lysine (K1) near the E2 active site [3].
- Engineering for Enhanced Efficiency: Structure-guided optimization mutated the C-terminal serine (S6) to glutamate, creating the superior acceptor tag KEGYEE. This sequence shows higher ubiquitination efficiency by UBE2E1 [3].
- Versatility: The SUE1 system can be used to generate monoubiquitinated proteins, diUb with defined linkages, polyUb chains, and even branched ubiquitin chains. Remarkably, it can also be adapted for the conjugation of the ubiquitin-like modifier NEDD8 [3].
Diagram 2: SUE1 mechanism for E3-independent ubiquitination.
Table 2: Key Ubiquitin Acceptor Tags and Their Properties
| Tag Name | Amino Acid Sequence | Recognizing Enzyme | Key Features & Applications |
|---|---|---|---|
| bioGEFUb(^nc) | AviTag (GEF mutant) fused to Ubiquitin (L73P) | BirA-E3 Fusions | Proximity biotinylation; identification of E3 substrates in cells [22]. |
| SUE1 Tag (Optimal) | KEGYEE | UBE2E1 E2 Enzyme | E3-free monoubiquitination; generation of defined polyUb and branched chains [3]. |
| SUE1 Tag (Native) | KEGYES | UBE2E1 E2 Enzyme | Native sequence for E3-free ubiquitination of SETDB1 [3]. |
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents for Engineered Ubiquitination Systems
| Reagent / Tool | Function / Description | Example Application |
|---|---|---|
| bioGEFUbnc | A non-cleavable, biotinylatable ubiquitin with a low-affinity AviTag variant. | Substrate identification via the BioE3 platform; minimizes non-specific labeling [22]. |
| BirA-E3 Fusion | A fusion protein combining a biotin ligase with an E3 ligase of interest. | Drives proximity-dependent biotinylation of ubiquitinated substrates in BioE3 [22]. |
| eSCR Fusion Protein | An engineered fusion of SKP1, Cullin1, and RBX1. | Core component for reconstituting modular, active SCF E3 ligases in vitro [23]. |
| UBE2E1 Enzyme | A unique human E2 conjugating enzyme. | Catalyzes E3-free, site-specific ubiquitination of the KEGY(E/D)(E/S) tag in the SUE1 system [3]. |
| K29/K48-branched triUb probe | A chemically synthesized ubiquitin probe mimicking a branched chain. | Used to trap enzymatic intermediates for structural studies (e.g., cryo-EM of Ufd4) [24]. |
| Boc-Lisdexamfetamine | Boc-Lisdexamfetamine|High-Purity Reference Standard | Boc-Lisdexamfetamine is a protected derivative for analytical research and method development. This product is for Research Use Only (RUO). Not for human consumption. |
| 4'-Thioguanosine | 4'-Thioguanosine | 4'-Thioguanosine is a nucleoside analog for anticancer and anti-HBV research. This product is for research use only and not for human consumption. |
Detailed Experimental Protocols
Protocol: BioE3 for Identifying E3 Ligase Substrates
Application: Identifying bona fide cellular substrates for a specific E3 ligase. Principle: A BirA-E3 fusion protein co-localizes with bioGEFUbnc during substrate ubiquitination, leading to proximity-based biotinylation of the substrate, which is then purified and identified.
Procedure:
- Generate Stable Cell Line: Create a U2OS or HEK293FT cell line with a DOX-inducible bioGEFUbnc construct.
- Culture and Deplete Biotin: Culture the cells in dialyzed, biotin-depleted serum for at least 24 hours prior to the experiment.
- Express BirA-E3 Fusion: Transfect the cells with the BirA-E3 fusion construct.
- Induce and Label: Add DOX to induce bioGEFUbnc expression and BirA-E3 fusion expression. After 24 hours, add exogenous biotin to the culture medium for a limited time window (e.g., 2 hours).
- Harvest and Lyse Cells: Harvest cells and lyse in a suitable buffer (e.g., RIPA) containing protease inhibitors and N-ethylmaleimide (NEM) to inhibit deubiquitinases.
- Streptavidin Pulldown: Incubate the clarified lysate with streptavidin-coated beads under stringent washing conditions to capture biotinylated proteins.
- Elution and Analysis: Elute the bound proteins and identify them using LC-MS/MS. Compare against appropriate negative controls (e.g., BirA alone) to filter out non-specific interactors [22].
Protocol: SUE1 for E3-Free Generation of Ubiquitinated Proteins
Application: In vitro generation of site-specifically ubiquitinated or neddylated proteins with customized chain architectures. Principle: The UBE2E1 enzyme directly recognizes the optimized KEGYEE tag on a substrate and transfers ubiquitin without the need for an E3 ligase.
Procedure:
- Clone and Express: Engineer the target protein to contain the KEGYEE sequence at the desired C-terminal or internal location. Express and purify the tagged substrate protein.
- Purify Enzymes: Purify the necessary enzymes: E1 (Uba1), E2 (UBE2E1), and ubiquitin (or NEDD8).
- Set Up Ubiquitination Reaction: Assemble a reaction mixture containing:
- 50 mM Tris-HCl, pH 7.5
- 5 mM MgClâ‚‚
- 2 mM ATP
- 0.1-1 µM E1 (Uba1)
- 1-5 µM E2 (UBE2E1)
- 50-100 µM Ubiquitin (or desired Ub mutant for specific linkages)
- 1-10 µM Substrate protein (with KEGYEE tag)
- Incubate and Analyze: Incubate the reaction at 30°C for 1-3 hours. Stop the reaction by adding SDS-PAGE loading buffer (without reducing agents if analyzing thioester intermediates). Analyze the products by immunoblotting or mass spectrometry to confirm site-specific modification [3].
The Ubiquiton system is an engineered set of ubiquitin protein ligases and matching ubiquitin acceptor tags designed to enable rapid, inducible, and linkage-specific polyubiquitylation of proteins of interest (POIs) in both yeast and mammalian cells [7]. This protocol provides a detailed methodology for implementing this groundbreaking tool to explore the diverse signaling functions of polyubiquitin chains in biological contexts ranging from proteasomal targeting to endocytic pathways [7] [15].
The Ubiquiton system addresses a significant limitation in ubiquitin research—the inability to experimentally induce specific polyubiquitin chain linkages on target proteins in cells [7]. By providing precise control over ubiquitin chain topology, researchers can now establish direct cause-effect relationships between specific ubiquitin modifications and their functional outcomes.
Principles of the Ubiquiton System
The Ubiquiton system operates through the induced interaction between engineered E3 ubiquitin ligases and ubiquitin acceptor tags fused to proteins of interest. This interaction triggers the formation of specific polyubiquitin chain types on the target protein.
Diagram 1: Ubiquiton System Mechanism. The system functions through inducible interaction between the engineered E3 ligase and ubiquitin acceptor tag, resulting in linkage-specific polyubiquitin chain formation.
Materials and Reagents
Key Research Reagent Solutions
Table 1: Essential Components of the Ubiquiton System
| Component | Type/Function | Specific Variants Available | Key Applications |
|---|---|---|---|
| Engineered E3 Ligases | Catalyze specific ubiquitin chain formation | Linear (M1-), K48-, K63-specific ligases | Inducing specific ubiquitin linkages |
| Ubiquitin Acceptor Tags | Fusion tags that receive ubiquitin chains | Matching tags for different E3 ligases | Targeting proteins of interest |
| Induction System | Controls E3-tag interaction | Rapamycin, ABA, or other inducible systems | Temporal control of ubiquitylation |
| Expression Vectors | Mammalian and yeast systems | Plasmids for POI-tag fusions | System implementation in different cell types |
| Detection Antibodies | Validate ubiquitin linkages | Linkage-specific ubiquitin antibodies | Confirmation of specific chain types |
Required Supporting Reagents
- Cell Lines: Appropriate mammalian (HEK293, U2OS, etc.) or yeast cells compatible with the chosen expression system
- Culture Media: Standard media with appropriate selection antibiotics
- Transfection Reagents: For plasmid delivery (e.g., lipofectamine, calcium phosphate, electroporation reagents)
- Induction Agents: Specific to the chosen induction system (e.g., rapamycin for dimerization systems)
- Lysis Buffers: RIPA or similar buffers with protease inhibitors and deubiquitinase inhibitors (N-ethylmaleimide)
- Immunoblotting Materials: SDS-PAGE gels, transfer membranes, and detection reagents
- Primary Antibodies: Anti-ubiquitin, linkage-specific ubiquitin antibodies, and antibodies against your protein of interest
- Secondary Antibodies: HRP or fluorescence-conjugated antibodies for detection
Experimental Protocol
Phase 1: System Design and Construct Preparation
Step 1.1: Select Appropriate Ubiquiton Components
Choose the specific E3 ligase and matching ubiquitin acceptor tag based on your desired ubiquitin linkage type and experimental system. The Ubiquiton system currently supports linear (M1-), K48-, and K63-linked polyubiquitylation [7].
Step 1.2: Clone Protein of Interest
Clone your gene of interest into the appropriate expression vector containing the ubiquitin acceptor tag using standard molecular biology techniques:
- Use restriction enzyme digestion and ligation or Gibson assembly
- Verify reading frame maintenance between POI and tag
- Sequence verify all constructs
Step 1.3: Prepare E3 Ligase Construct
Prepare the engineered E3 ligase expression vector matching your chosen ubiquitin acceptor tag.
Phase 2: Cell Culture and Transfection
Step 2.1: Cell Seeding
- Seed appropriate cells in 6-well or 12-well plates at 30-50% confluence
- Allow cells to adhere overnight under standard culture conditions
Step 2.2: Transfection
Transfert cells with both POI-tag fusion and engineered E3 ligase constructs:
- DNA Ratios: Optimize DNA ratios (typically 1:1 POI-tag:E3 ligase)
- Transfection Method: Use preferred method (lipofection, calcium phosphate, etc.)
- Controls: Include empty vector and tag-only controls
Step 2.3: Post-Transfection Incubation
Incubate cells for 24-48 hours to allow protein expression before induction.
Phase 3: Induction of Ubiquitylation
Step 3.1: Induction Agent Application
Apply the appropriate induction agent to trigger E3 ligase-ubiquitin acceptor tag interaction:
- Timing: Typically 1-6 hours before harvesting
- Concentration: Optimize for your system (e.g., 1-100 nM rapamycin)
- Controls: Include non-induced controls for comparison
Step 3.2: Monitor Induction
Monitor cells for potential toxicity effects and morphological changes during induction.
Phase 4: Sample Processing and Analysis
Step 4.1: Cell Harvesting and Lysis
- Wash cells with ice-cold PBS
- Lyse cells in appropriate buffer containing protease and deubiquitinase inhibitors
- Clarify lysates by centrifugation (14,000 × g, 15 minutes, 4°C)
Step 4.2: Ubiquitylation Detection
Analyze ubiquitylation using multiple complementary methods:
Method A: Immunoblotting
- Separate proteins by SDS-PAGE (6-12% gradient gels recommended)
- Transfer to PVDF or nitrocellulose membranes
- Probe with anti-ubiquitin and protein-specific antibodies
- Use linkage-specific ubiquitin antibodies to verify chain type
Method B: Immunoprecipitation
- Immunoprecipitate POI-tag fusion under denaturing conditions
- Analyze precipitates by immunoblotting with ubiquitin antibodies
Method C: Mass Spectrometry
- For advanced verification, use mass spectrometry to confirm linkage specificity
Data Interpretation and Validation
Expected Results and Controls
Table 2: Expected Outcomes and Validation Criteria for Ubiquiton Experiments
| Experimental Condition | Expected Ubiquitylation Pattern | Validation Approach | Common Pitfalls |
|---|---|---|---|
| Complete system + induction | Strong, linkage-specific polyubiquitin smearing | Shift in molecular weight; linkage-specific antibody recognition | Non-specific background |
| E3 ligase only + induction | No POI ubiquitylation | Absence of high molecular weight species | Non-specific E3 activity |
| POI-tag only + induction | No POI ubiquitylation | Absence of high molecular weight species | Endogenous ubiquitylation |
| Complete system - induction | Minimal background ubiquitylation | Low basal ubiquitylation levels | Leaky induction system |
| Different linkage systems | Distinct functional outcomes | Functional assays specific to chain type | Cross-reactivity between systems |
Functional Validation Experiments
The Ubiquiton system has been validated for multiple cellular applications:
For K48-Linked Chains:
- Monitor protein degradation via cycloheximide chase assays
- Assess proteasomal dependence using MG132 or other proteasome inhibitors
For K63-Linked Chains:
- Examine endocytic trafficking through microscopy or surface biotinylation
- Monitor protein-protein interactions by co-immunoprecipitation
For Linear (M1) Linked Chains:
- Assess NF-κB signaling through reporter assays or target gene analysis
Troubleshooting Guide
Table 3: Common Technical Issues and Resolution Strategies
| Problem | Potential Causes | Solutions | Preventive Measures |
|---|---|---|---|
| No ubiquitylation detected | Poor transfection efficiency, incorrect construct design | Optimize transfection, verify constructs | Include positive control constructs |
| High background ubiquitylation | Leaky induction system, non-specific E3 activity | Adjust induction system, use tighter promoters | System optimization in control cells |
| Incorrect chain linkage | Off-target E3 activity, antibody cross-reactivity | Verify linkage specificity with multiple methods | Include appropriate linkage controls |
| Cellular toxicity | Overexpression effects, excessive ubiquitylation | Titrate expression levels, shorten induction time | Time-course and dose-response experiments |
| Poor detection sensitivity | Low expression, inefficient ubiquitylation | Enhance expression, improve detection methods | Optimize protein stability and detection |
Applications and Extended Protocols
The Ubiquiton system has been successfully applied to diverse biological contexts:
Protocol 1: Controlling Protein Stability via K48-Linked Ubiquitylation
Specialized Materials:
- Proteasome inhibitors (MG132, bortezomib)
- Protein synthesis inhibitors (cycloheximide, anisomycin)
Procedure:
- Introduce POI with K48-specific Ubiquiton system
- Induce ubiquitylation for predetermined time
- Treat with cycloheximide to block new protein synthesis
- Harvest cells at time points (0, 30, 60, 120, 240 minutes)
- Analyze POI levels by immunoblotting, quantifying band intensity
Data Interpretation:
- Compare degradation rates between induced and non-induced conditions
- Verify proteasomal dependence with MG132 co-treatment
Protocol 2: Modulating Endocytic Trafficking via K63-Linked Ubiquitylation
Specialized Materials:
- Surface biotinylation reagents (NHS-SS-biotin, streptavidin beads)
- Immunofluorescence reagents for microscopy
Procedure:
- Express membrane protein with K63-specific Ubiquiton system
- Induce ubiquitylation for optimized time
- Perform surface biotinylation at 4°C
- Warm cells to 37°C for various times to allow internalization
- Strip remaining surface biotin and quantify internalized protein
Data Interpretation:
- Accelerated internalization expected with K63-linked ubiquitylation
- Confirm by immunofluorescence showing altered localization
Advanced Experimental Design
Timing Considerations
- Acute vs. Chronic Induction: Short induction (1-2 hours) for acute effects; longer induction (4-24 hours) for cumulative effects
- Degradation Kinetics: Optimize time points based on protein half-life
- Functional Outcomes: Align ubiquitylation induction with appropriate assay timelines
System Extensions
- Combination with Other Tools: Use alongside CRISPR/Cas9, degron technologies, or biosensors
- Multiplexed Approaches: Combine multiple Ubiquiton systems to study competing ubiquitin signals
- Spatiotemporal Control: Implement with light-inducible or compartment-specific systems
Diagram 2: Comprehensive Ubiquiton Workflow. The complete experimental pathway from system design to functional analysis, demonstrating the versatility of the Ubiquiton platform.
The Ubiquiton system represents a transformative approach for investigating linkage-specific ubiquitin signaling. This detailed protocol enables researchers to implement this technology across diverse biological contexts, from controlling protein stability to modulating cellular trafficking and signaling pathways. The inducible nature of the system provides temporal control that is essential for establishing causal relationships between specific ubiquitin modifications and their functional consequences, advancing our understanding of the ubiquitin code in health and disease.
Controlling Protein Localization and Stability with Ubiquiton
The post-translational modification of proteins by ubiquitin is a fundamental regulatory mechanism that controls an enormous range of physiological processes, including protein degradation, membrane trafficking, and signal transduction [25]. The versatility of ubiquitin signaling stems from its ability to form polymeric chains through different lysine linkages, with each linkage type encoding distinct functional consequences for the modified protein [7]. For decades, researchers have lacked experimental tools to precisely induce specific polyubiquitin chain types on proteins of interest in live cells, limiting our ability to decipher the ubiquitin code.
The Ubiquiton system represents a breakthrough in ubiquitin research methodology, providing a set of engineered ubiquitin protein ligases and matching ubiquitin acceptor tags for rapid, inducible, and linkage-specific polyubiquitylation of target proteins [7] [26]. This innovative tool enables unprecedented control over protein localization and stability, allowing researchers to directly manipulate cellular processes through targeted ubiquitin signaling.
The Ubiquiton System: Components and Mechanism
System Architecture
The Ubiquiton system consists of two core components: engineered E3 ubiquitin ligases and matching ubiquitin acceptor tags. These components work in concert to achieve linkage-specific polyubiquitylation of target proteins:
- Engineered E3 Ligases: Specifically designed to synthesize linear (M1-), K48-, or K63-linked polyubiquitin chains
- Ubiquitin Acceptor Tags: Genetically fused to proteins of interest, serving as optimized substrates for the engineered ligases
- Inducible Design: Enables temporal control over the ubiquitination process [7]
Molecular Mechanism
The system hijacks the natural ubiquitination cascade while introducing specificity and control:
- Activation: Ubiquitin is activated by E1 enzymes in an ATP-dependent process
- Conjugation: Activated ubiquitin is transferred to E2 conjugating enzymes
- Ligation: Engineered E3 ligases specifically transfer ubiquitin to acceptor tags, building chains with defined linkages [25]
This mechanism enables the formation of ubiquitin chains with precise connectivity, overcoming the natural promiscuity of endogenous ubiquitination machinery.
Applications and Experimental Validation
Functional Applications
The Ubiquiton system has been validated across multiple biological contexts and protein types:
| Application Domain | Validated Function | Ubiquitin Linkage |
|---|---|---|
| Proteasomal Targeting | Induces degradation of soluble proteins | K48-linked |
| Membrane Protein Trafficking | Controls endocytosis and localization | K63-linked |
| Chromatin-associated Proteins | Regulates DNA-related processes | Linear (M1-linked) |
| Nuclear Proteins | Modulates transcription factor activity | Multiple linkage types |
Researchers have successfully applied Ubiquiton to soluble cytoplasmic proteins, nuclear proteins, chromatin-associated factors, and integral membrane proteins, demonstrating its broad utility [7].
Quantitative Assessment
Experimental data from Ubiquiton applications reveal key performance metrics:
| Parameter | Performance | Experimental Context |
|---|---|---|
| Temporal Control | Rapid induction (minutes) | Yeast and mammalian cells |
| Specificity | Exclusive formation of designated linkage | In vitro and in vivo |
| Functional Impact | Efficient protein degradation or relocalization | Multiple substrate types |
| Versatility | Compatible with diverse protein classes | Cytosolic, nuclear, membrane proteins |
Experimental Protocols
Protocol 1: Induction of Linkage-Specific Polyubiquitylation in Cells
This protocol enables researchers to induce specific polyubiquitin chain formation on target proteins in live cells.
Materials and Reagents
- Cells expressing Ubiquiton components (engineered E3 ligase and tagged target protein)
- Induction agent (varies by specific Ubiquiton variant)
- Lysis buffer: 50 mM HEPES (pH 8.0), 150 mM NaCl, 1% Triton X-100, protease inhibitors
- SDS-PAGE and Western blot equipment
- Ubiquitin linkage-specific antibodies
Procedure
- Cell Preparation: Culture cells expressing both the engineered E3 ligase and the target protein fused to the ubiquitin acceptor tag
- Induction: Apply induction agent according to optimized kinetics (typically 30-120 minutes)
- Harvesting: Collect cells by centrifugation and wash with PBS
- Lysis: Resuspend cell pellet in ice-cold lysis buffer and incubate for 15 minutes on ice
- Clarification: Centrifuge at 15,000 × g for 15 minutes at 4°C
- Analysis:
- Separate proteins by SDS-PAGE
- Transfer to membrane and probe with ubiquitin linkage-specific antibodies
- Reprobe with target protein antibody to confirm modification
Troubleshooting
- Low Ubiquitination Efficiency: Optimize expression levels of E3 and target protein
- Basal Activity Before Induction: Titrate expression levels to minimize leakiness
- Non-specific Ubiquitination: Include controls with catalytically inactive E3 variants
Protocol 2: Functional Assessment of Ubiquitinated Proteins
This protocol outlines methods to evaluate the functional consequences of targeted ubiquitination.
Protein Stability Assay
- Induce polyubiquitylation as described in Protocol 1
- Block new protein synthesis using cycloheximide (100 µg/mL)
- Collect samples at time points (0, 15, 30, 60, 120 minutes)
- Analyze target protein levels by Western blotting
- Quantify band intensity and calculate half-life
Protein Localization Assay
- Culture cells expressing Ubiquiton components on glass coverslips
- Induce polyubiquitylation
- Fix cells with 4% paraformaldehyde at various time points
- Permeabilize with 0.1% Triton X-100
- Stain with target protein antibody and appropriate fluorescent secondary
- Image using confocal microscopy
- Quantify localization changes using image analysis software
Research Reagent Solutions
Essential materials and reagents for implementing Ubiquiton technology:
| Reagent Category | Specific Examples | Function/Purpose |
|---|---|---|
| Engineered E3 Ligases | M1-specific, K48-specific, K63-specific ligases | Catalyze formation of specific ubiquitin linkages |
| Ubiquitin Acceptor Tags | Linear, K48, K63 acceptor variants | Serve as optimized substrates for polyubiquitylation |
| Activation Enzymes | E1 ubiquitin-activating enzyme | Initiates ubiquitin activation in ATP-dependent manner |
| Conjugation Enzymes | E2 ubiquitin-conjugating enzymes | Transfers activated ubiquitin to E3 ligases |
| Detection Reagents | Linkage-specific ubiquitin antibodies | Enable detection of specific polyubiquitin chains |
| Cell Lines | Yeast and mammalian expression systems | Provide cellular context for Ubiquiton application |
Schematic Representations
Ubiquiton System Mechanism
Experimental Workflow for Protein Degradation Analysis
The Ubiquiton system represents a transformative tool for precise manipulation of protein fate in research and drug development. By enabling inducible, linkage-specific polyubiquitylation, this technology provides unprecedented control over protein stability and localization, opening new avenues for investigating ubiquitin-dependent processes and developing targeted protein degradation therapies. As the ubiquitin field continues to evolve, Ubiquiton stands as a versatile platform for deciphering the complex language of ubiquitin signaling in health and disease.
This application note details the use of the Ubiquiton system, an inducible, linkage-specific polyubiquitylation tool, for controlling diverse cellular processes by directing specific ubiquitin signals to proteins of interest across various cellular compartments [7] [15]. We provide validated protocols and reagent information to enable researchers to deploy this tool effectively.
The Ubiquiton system addresses a critical gap in ubiquitin research by enabling the rapid, inducible, and linkage-specific polyubiquitylation of target proteins in live cells [7]. It consists of a modular pair of components:
- Engineered E3 Ubiquitin Ligases: Designed to synthesize only one specific type of polyubiquitin chain (linear/M1-, K48-, or K63-linked) [7].
- Matching Ubiquitin Acceptor Tags: Short peptide tags (degrons) that are fused to the protein of interest and serve as a defined substrate for the engineered E3 ligases [7].
This system allows precise control over the ubiquitin code, moving beyond simple degradation to explore the diverse signaling functions of polyubiquitin chains in regulating protein localization, activity, and complex formation [7] [27].
The Ubiquiton system has been quantitatively validated for a range of targets and cellular processes. The table below summarizes key experimental findings.
Table 1: Summary of Ubiquiton System Applications and Quantitative Outcomes
| Target Protein / Process | Cellular Location | Ubiquitin Linkage Induced | Key Functional Outcome | Quantitative/Experimental Readout |
|---|---|---|---|---|
| Proteasomal Targeting [7] | Cytoplasm & Nucleus | K48-linked | Induced degradation of soluble proteins [7] | Rapid loss of target protein observed via immunoblotting [7] |
| Endocytic Pathway [7] | Plasma Membrane | K63-linked & Monoubiquitylation | Controlled internalization of membrane proteins (e.g., EGFR) [7] | Altered protein localization and stability confirmed by imaging and biochemical methods [7] |
| Ion Channel Trafficking (KCNQ1) [27] | Plasma Membrane & Intracellular Compartments | N/A (Analysis of endogenous chains) | Defined roles for endogenous chains: K48 for forward trafficking; K63 for enhanced endocytosis [27] | Flow cytometry for surface expression; confocal microscopy for colocalization; mass spectrometry identified dominant K48 (72%) and K63 (24%) chains [27] |
| Synaptic Protein Regulation [28] | Postsynaptic Density & Presynaptic Terminals | K63-linked | Regulated synaptic content and function, independent of proteasomal degradation [28] | Immunoblotting showed increased K63-polyUb levels; proteomic analysis identified ubiquitinated pre- and postsynaptic proteins [28] |
Experimental Protocols
Protocol: Inducing Linkage-Specific Protein Degradation
This protocol describes using the Ubiquiton system to induce degradation of a cytosolic or nuclear protein of interest (POI) via K48-linked polyubiquitylation [7].
Key Research Reagent Solutions:
- Plasmids: Engineered E3 ligase for K48-linked chains; expression vector for POI fused to a matching ubiquitin acceptor tag (degron) [7].
- Inducer: Doxycycline or other suitable agent, depending on the system configuration [7].
- Antibodies: Anti-FLAG M2 antibody (for detection of tagged proteins) [9]. Anti-Ubiquitin antibody, Lys48-Specific (clone Apu3) to confirm chain linkage [9].
- Inhibitors: Cycloheximide (to block new protein synthesis and monitor degradation kinetics) [9]. MG132 (proteasome inhibitor) for control experiments [27].
Methodology:
- Cell Seeding and Transfection: Seed HEK293 or other suitable mammalian cells in a 6-well plate. The next day, co-transfect cells with two plasmids: one expressing the K48-specific engineered E3 ligase and another expressing your POI fused to the matching degron tag.
- Induction of Ubiquitylation: 24 hours post-transfection, induce the expression of the E3 ligase by adding doxycycline (e.g., 1 µg/mL) to the culture medium. To monitor degradation kinetics, add cycloheximide (e.g., 100 µg/mL) to inhibit new protein synthesis.
- Sample Collection: Lyse cells at various time points post-induction (e.g., 0, 1, 2, 4 hours) in RIPA buffer supplemented with protease inhibitors (e.g., SIGMAFAST Protease Inhibitor Cocktail Tablets) and 10 mM N-ethylmaleimide (NEM) to preserve ubiquitin conjugates [9].
- Analysis:
- Immunoblotting: Resolve cell lysates by SDS-PAGE and transfer to a membrane. Probe with an antibody against your POI or its tag (e.g., anti-FLAG) to monitor protein degradation over time.
- Linkage Specificity Confirmation: Perform immunoprecipitation of the POI under denaturing conditions. Probe the blot with a K48-linkage specific ubiquitin antibody (e.g., clone Apu3) to confirm the formation of K48-linked chains [9].
Protocol: Controlling Membrane Protein Localization via K63-Linked Ubiquitylation
This protocol outlines the use of the Ubiquiton system to induce K63-linked ubiquitylation on a membrane protein, such as EGFR, to control its endocytosis and endosomal sorting [7].
Key Research Reagent Solutions:
- Plasmids: Engineered E3 ligase for K63-linked chains; expression vector for membrane POI (e.g., EGFR) fused to a matching ubiquitin acceptor tag [7].
- Antibodies: Anti-Ubiquitin antibody, Lys63-Specific (for confirming chain linkage). Anti-VSV Glycoprotein antibody (clone P5D4) if using a VSV-tagged protein [9].
- Other Reagents: Chloroquine (lysosome inhibitor) for control experiments [9]. Cell surface protein biotinylation kit.
Methodology:
- Cell Preparation and Transfection: Seed cells appropriate for your membrane protein (e.g., HeLa). Co-transfect with the K63-specific E3 ligase and the membrane POI-degron construct.
- Induction and Stimulation: Induce E3 ligase expression with doxycycline. If studying a receptor like EGFR, serum-starve cells and then stimulate with EGF to activate the receptor and initiate trafficking.
- Monitoring Internalization:
- Surface Biotinylation: At various time points post-induction/stimulation, label cell surface proteins with a non-cell-permeant biotin reagent. Quench the reaction, lyse cells, and pull down biotinylated proteins with streptavidin beads. Detect your POI by immunoblotting to quantify the amount remaining at the surface.
- Immunofluorescence Microscopy: Fix cells and stain for the POI and early endosome markers (e.g., EEA1). Use confocal microscopy to quantify the co-localization of the POI with endosomal compartments, demonstrating its ligand-induced internalization, which is enhanced by K63-linked ubiquitylation.
- Validation: Immunoprecipitate the POI and probe for ubiquitin with a K63-linkage specific antibody to confirm the specific modification [7].
The Scientist's Toolkit: Essential Research Reagents
The following table catalogs key reagents used in Ubiquiton system experiments, as evidenced by the cited studies.
Table 2: Key Research Reagent Solutions for Ubiquiton Experiments
| Reagent / Material | Function / Application | Specific Examples / Clones |
|---|---|---|
| Linkage-Specific Ubiquitin Antibodies | Critical for validating the specific type of polyubiquitin chain assembled on the target protein. | Anti-Ubiquitin, Lys63-Specific (clone Apu3); Anti-Linear Ubiquitin (clone LUB9) [9]. |
| General Ubiquitin Detection Antibody | Detects total ubiquitin conjugates, useful for initial confirmation of ubiquitylation. | Anti-Ubiquitinylated proteins (clone FK2) [9]. |
| Epitope Tag Antibodies | For detecting and immunoprecipitating tagged target proteins and components of the Ubiquiton system. | ANTI-FLAG M2 antibody; Anti-VSV Glycoprotein (clone P5D4); Anti-GFP [9]. |
| Proteasome Inhibitor | Used in control experiments to confirm that a phenotypic outcome (e.g., loss of protein) is proteasome-dependent. | MG132 [27]. |
| Lysosome Inhibitor | Used in control experiments to determine if a protein is degraded via the endolysosomal pathway. | Chloroquine diphosphate [9]. |
| Protein Synthesis Inhibitor | Used in degradation kinetic experiments to block new protein synthesis, allowing clear monitoring of protein turnover. | Cycloheximide [9]. |
| Protease Inhibitor Cocktail | Essential for preserving protein integrity and post-translational modifications during cell lysis and protein extraction. | SIGMAFAST Protease Inhibitor Cocktail Tablets, EDTA-Free [9]. |
| Dodemorph benzoate | Dodemorph Benzoate|CAS 59145-63-0|RUO | Dodemorph benzoate is a morpholine fungicide for powdery mildew research. This product is For Research Use Only and is not intended for personal use. |
| 1-Benzyl-2-naphthol | 1-Benzyl-2-naphthol | 1-Benzyl-2-naphthol is a key C-alkylated product and Betti base precursor for asymmetric synthesis research. This product is for research use only (RUO). Not for human use. |
Maximizing Experimental Success: Troubleshooting and Optimizing Ubiquiton
The Ubiquiton inducible, linkage-specific polyubiquitylation tool represents a significant advancement in the synthetic control of cellular signaling [15]. This technology enables researchers to precisely induce the attachment of polyubiquitin chains with defined linkage types to target proteins of interest, allowing for the dissection of the diverse functional outcomes signaled by different ubiquitin topologies. Central to this system is the efficient induction of the ubiquitination machinery and the subsequent accessibility of the engineered tags.
Simultaneously, the development of ubi-tagging showcases a complementary, powerful methodology for protein engineering. Ubi-tagging is a modular and versatile technique for the site-directed multivalent conjugation of antibodies, antibody fragments, nanobodies, peptides, or small molecules via ubiquitin [29]. It leverages the native ubiquitination enzymatic cascade—comprising ubiquitin-activating (E1), ubiquitin-conjugating (E2), and ubiquitin-ligating (E3) enzymes—to create defined protein conjugates rapidly, often within 30 minutes. The successful application of both the Ubiquiton tool and ubi-tagging methodologies is critically dependent on two fundamental parameters: achieving high induction efficiency of the system components and ensuring the unhindered accessibility of the ubiquitin tags for enzymatic processing. Failures in these areas represent the most common pitfalls encountered during experimental deployment.
Critical Pitfalls and Optimization Strategies
The following table summarizes the primary pitfalls related to induction efficiency and tag accessibility, along with their experimental consequences and optimized solutions.
Table 1: Common Pitfalls in Induction Efficiency and Tag Accessibility
| Pitfall Category | Specific Pitfall | Impact on Experiment | Optimized Solution |
|---|---|---|---|
| Induction Efficiency | Suboptimal expression of E3 ligase or Ubiquiton construct | Low ubiquitylation yield; high background; failed experiments | Titrate inducer concentration; use high-efficiency expression vectors (e.g., CRISPR/HDR [29]); verify expression via Western blot |
| Induction Efficiency | Inefficient E2–E3 Enzyme Pairing | Poor conjugation efficiency and slow reaction kinetics | Use fusion proteins of E2 and E3 (e.g., gp78RING-Ube2g2 for K48 linkages [29]); pre-validate enzyme activity |
| Tag Accessibility | Steric Hindrance from Large Protein Cargo | Reduced conjugation efficiency; incomplete product formation | Use flexible peptide linkers between the tag and cargo; consider smaller protein tags or fragments |
| Tag Accessibility | Incorrect Ubi-Tag Design (Donor/Acceptor) | Uncontrolled homodimerization or polymerization; heterogeneous products | Use designated donor (Ubdon-K48R) and acceptor (Ubacc-ΔGG) tags [29] |
| Reaction Conditions | Non-physiological Buffer Components | Compromised enzyme activity and protein stability | Maintain physiological pH; avoid amine-containing buffers (e.g., Tris) in conjugation reactions [30] |
| Reaction Conditions | Presence of Aggregation-Prone Sequences | Protein aggregation, loss of functional product, and low recovery | Include stabilizing agents; optimize purification to remove aggregates (aim for <5% aggregation [31]) |
Essential Protocols for System Validation
Protocol 1: Quantitative Assessment of Induction Efficiency
This protocol outlines the steps to quantify the expression levels of key Ubiquiton system components (e.g., the inducible E3 ligase) following induction.
- Induction and Lysis: Induce your expression system (e.g., with doxycycline or other inducers) in a controlled manner. Harvest cells and lyse them using a non-denaturing RIPA buffer supplemented with proteasome inhibitors (e.g., MG132) and deubiquitinase (DUB) inhibitors (e.g., PR-619) to preserve ubiquitination states.
- Protein Quantification and Separation: Determine the total protein concentration of the lysates using a Bradford or BCA assay. Separate equal amounts of protein (e.g., 20-30 µg) by SDS-PAGE.
- Immunoblotting: Transfer proteins to a PVDF membrane and probe with antibodies specific against your tagged protein (e.g., anti-FLAG for the Ubiquiton construct) and a loading control (e.g., GAPDH).
- Densitometric Analysis: Use image analysis software (e.g., ImageJ) to perform densitometry on the immunoblot bands. Calculate the induction efficiency as the ratio of the signal intensity of the induced sample to that of the uninduced control, normalized to the loading control. An efficient induction should yield a >10-fold increase in expression.
Protocol 2: Functional Validation of Ubi-Tag Accessibility via Conjugation
This protocol describes a functional assay to confirm that the ubi-tag is accessible for enzymatic conjugation, based on established ubi-tagging workflows [29].
- Reaction Setup: In a final volume of 50 µL of conjugation buffer (e.g., PBS, pH 7.4), combine the following components:
- Purified Ubi-tagged protein of interest (e.g., Fab-Ub(K48R)don): 10 µM
- Acceptor tag (e.g., Rho-Ubacc-ΔGG): 50 µM (5-fold excess)
- Recombinant E1 enzyme: 0.25 µM
- Linkage-specific E2–E3 fusion enzyme (e.g., gp78RING-Ube2g2 for K48): 20 µM
- ATP (as a 10x solution in MgClâ‚‚): 1x final concentration
- Incubation and Termination: Incubate the reaction mixture at 37°C for 30 minutes. Stop the reaction by adding 10 µL of 6x SDS-PAGE loading buffer (without reducing agents if checking for conjugate formation).
- Analysis:
- SDS-PAGE: Analyze 10-15 µL of the reaction product by non-reducing SDS-PAGE. Successful conjugation is indicated by an upward band shift on a Coomassie-stained gel.
- Fluorescence Detection: If using a fluorescent acceptor (e.g., rhodamine), visualize the gel using a fluorescence imager to confirm the conjugation of the payload to the target protein.
- Mass Spectrometry: For precise validation, purify the conjugate and analyze it by Electrospray Ionization Time-of-Flight (ESI-TOF) mass spectrometry. The observed mass should correspond to the calculated mass of the conjugate, with complete consumption of the starting ubi-tagged protein [29].
The Scientist's Toolkit: Key Research Reagent Solutions
The following table catalogs essential reagents and kits for ubiquitination and protein conjugation research, providing researchers with a curated list of tools for their experiments.
Table 2: Essential Research Reagent Solutions for Ubiquitination and Conjugation Studies
| Reagent/Kits | Primary Function | Key Features and Applications |
|---|---|---|
| Ubi-Tagging Enzyme System | Site-specific protein conjugation | Recombinant E1 and linkage-specific E2-E3 fusion enzymes (e.g., gp78RING-Ube2g2); used for generating defined antibody conjugates and multispecific proteins [29]. |
| AGLink Site-Specific Conjugation Kit | Antibody-drug conjugation | Glycan-mediated conjugation at Fc N297; achieves DAR2 or DAR4 with high homogeneity (>90% efficiency); ideal for ADC screening and performance studies [31]. |
| Lightning-Link Conjugation Kits | Rapid antibody labeling | Labels antibodies with >45 labels (fluorophores, enzymes, proteins) in <4 hours with 30s hands-on time; targets primary amines; no purification step needed [30]. |
| Synthetic Ubiquitin Derivatives | Custom conjugation payloads | Chemically synthesized ubiquitin mutants (e.g., Ubacc-ΔGG) with N-terminal modifications (fluorophores, antigenic peptides); enables modular conjugate assembly [29]. |
| His-Tagged Ubiquitin Plasmids | Ubiquitylome profiling | Enables enrichment of ubiquitinated proteins from cell lysates for identification by LC-MS/MS; critical for substrate discovery [32]. |
| Extensumside H | Extensumside H|C21 Pregnane Glycoside|RUO | Extensumside H is a natural C21 pregnane glycoside for research on sweet taste receptors. For Research Use Only. Not for human or veterinary use. |
Workflow and Pathway Visualizations
Ubi-tagging Conjugation Workflow
The following diagram illustrates the core mechanism and workflow for generating defined protein conjugates using the ubi-tagging methodology.
Ubi-tagging Conjugation Workflow
Ubiquiton Inducible Polyubiquitylation Pathway
This diagram outlines the synthetic pathway for inducible, linkage-specific polyubiquitylation using the Ubiquiton tool, highlighting the critical control points.
Ubiquiton Inducible Polyubiquitylation Pathway
Optimization Strategies for Challenging Protein Targets
Targeted protein degradation (TPD) represents a paradigm shift in therapeutic development, enabling the direct targeting of proteins previously considered "undruggable." This application note details advanced optimization strategies for challenging protein targets, with a specific focus on the context of Ubiquiton inducible linkage-specific polyubiquitylation tools. We present structured experimental protocols, quantitative comparisons of key E3 ligases, and visualization of critical pathways to accelerate research in this rapidly evolving field. The integration of computational and experimental approaches provides a robust framework for developing next-generation degraders with enhanced precision and efficacy, addressing fundamental challenges in stability, bioavailability, and specificity that have constrained previous therapeutic modalities [33].
The ubiquitin-proteasome system (UPS) constitutes a sophisticated regulatory mechanism for controlling protein stability and function through covalent attachment of ubiquitin chains. Ubiquitination involves a sequential cascade mediated by ubiquitin-activating (E1), conjugating (E2), and ligase (E3) enzymes, with E3 ligases conferring substrate specificity [34]. The emergence of TPD technologies, particularly proteolysis-targeting chimeras (PROTACs) and molecular glues, has revolutionized drug discovery by leveraging this natural protein homeostasis machinery to selectively degrade pathological targets [33].
Despite this promise, critical challenges persist in optimizing TPD for challenging targets, including but not limited to: limited ligandable E3 ligases, suboptimal ubiquitin chain linkage specificity, restricted tissue penetration, and inadequate catalytic efficiency. The Ubiquiton inducible system addresses these limitations by enabling precise temporal and spatial control over ubiquitin chain formation, thereby facilitating the degradation of recalcitrant targets through customized polyubiquitin signatures [33] [35]. This application note provides a comprehensive methodological framework for leveraging these tools to overcome persistent bottlenecks in protein degradation.
Quantitative Analysis of E3 Ligase Properties
Table 1: Comparative Characteristics of Therapeutic E3 Ligases
| E3 Ligase | Domain Architecture | Linkage Specificity | Cellular Function | Therapeutic Relevance |
|---|---|---|---|---|
| Nedd4-2 | C2-WW1-WW2-WW3-WW4-HECT | K63-linked chains [36] | Regulates ion channels & membrane transporters; maintains Na+ homeostasis [36] | Hypertension, epilepsy, kidney diseases [36] |
| RNF114 | RING-Di19-UIM-Di19-UIM | K11-linked chains [35] | Extends K11 polyUb on MARUbylated substrates; DNA damage response [35] | Cancer, immune signaling pathways [35] |
| ZNF598 | RING-helical domain | Not specified | Resolves ribosome collisions at translation start sites [37] | Stem cell maintenance, neuronal function [37] |
| DTX2 | WWE-DTC-RING | Ub-ADPr ester linkage [35] | Generates initial MARUbe on PARP7 [35] | Cancer, interferon signaling [35] |
Table 2: E3 Ligase Optimization Parameters for Targeted Degradation
| Parameter | Optimization Strategy | Experimental Assessment | Success Metrics |
|---|---|---|---|
| Linkage Specificity | Engineer E2-E3 pairing interfaces; utilize Ubiquiton system with specified ubiquitin mutants | Mass spectrometry of ubiquitin chains; linkage-specific antibodies [35] | >80% desired linkage formation; minimal off-target chains |
| Catalytic Efficiency | Disrupt autoinhibitory domains; enhance E2 binding affinity | Fluorescence-based ubiquitination assays; autoubiquitination assays [36] | 3-5 fold increase in substrate turnover |
| Substrate Selectivity | Design multi-domain recruiters; optimize binding interfaces | Co-immunoprecipitation; proximity ligation assays [33] | <10% off-target binding; >90% target engagement |
| Tissue Penetration | Modify lipophilicity; implement transporter-mediated delivery | Brain-plasma ratio measurements; tissue homogenate analysis [33] | CNS access with >0.5 brain-plasma ratio |
Experimental Protocols
Protocol 1: Structural Characterization of E3 Auto-inhibition
Background: Many E3 ligases, including Nedd4-2, exist in autoinhibited states that limit their catalytic activity. Understanding these structural mechanisms enables rational activation for enhanced degradation efficiency [36].
Materials:
- Recombinantly expressed full-length E3 ligase (Nedd4-2 isoform 5, UniProt ID Q96PU5-5)
- Cryo-EM grids (Quantifoil R1.2/1.3, 300 mesh)
- 3 mM CHAPSO detergent
- Hydrogen/Deuterium Exchange (HDX-MS) buffers
- SV-AUC instrumentation (Beckman Coulter Optima AUC)
Methodology:
- Sample Preparation: Express and purify full-length Nedd4-2 using baculovirus system. Confirm phosphorylation status at S342 and S428 via LC-MS analysis for 14-3-3 binding studies [36].
- Cryo-EM Grid Preparation: Apply 3.5 μL of 0.5 mg/mL Nedd4-2 to glow-discharged grids. Blot for 3.5 seconds at 100% humidity and plunge-freeze in liquid ethane.
- Data Collection: Collect movies at 300 keV with defocus range of -0.8 to -2.2 μm. Process data through motion correction, CTF estimation, and particle picking (cryoSPARC).
- Structural Analysis: Reconstruct 3D volumes at 3.58-4.11 Ã… resolution. Identify C2 domain blocking E2-binding surface and WW1 domain masking ubiquitin-binding exosite [36].
- Functional Validation: Assess conformational changes induced by Ca²⺠(1 mM) through HDX-MS, monitoring deuterium incorporation in C2-HECT interface regions.
Troubleshooting: For particle heterogeneity, implement 3D variability analysis to separate conformational states. If resolution is limited at HECT domain, focus classification on this region.
Protocol 2: Optimization of Linkage-Specific Ubiquitination
Background: The Ubiquiton system enables precise control over ubiquitin chain topology. This protocol details optimization of K11-linked polyubiquitination using RNF114, which recognizes MARUbylated substrates through its Di19-UIM modules [35].
Materials:
- Fluorescent Ub-ADPr probe (synthesized via chemoenzymatic approach)
- RNF114 wild-type and mutant constructs
- MARUbylated PARP7 substrate
- E1 (UBA1), E2 (UBE2S), E3 (RNF114) enzymes
- ATP regeneration system
Methodology:
- MARUbe Recognition Assay: Incubate RNF114 (100 nM) with fluorescent Ub-ADPr probe (50 nM) in binding buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM DTT) for 30 minutes at 4°C.
- Complex Formation: Analyze RNF114-MARUbe interaction via AlphaFold3 modeling of the tandem Di19-UIM module (MARUbe-binding domain).
- Ubiquitination Reaction: Combine E1 (50 nM), E2 (UBE2S, 200 nM), RNF114 (500 nM), MARUbylated PARP7 (1 μM) in reaction buffer with ATP (2 mM). Incubate at 30°C for 60 minutes.
- Chain Analysis: Quench reaction with SDS sample buffer, resolve by SDS-PAGE, and immunoblot with K11-linkage specific antibody.
- Functional Validation: Conduct cellular knockdown experiments (siRNA against DTX2, RNF114) to confirm PARP7 MARUbylation dependency on both E3s.
Troubleshooting: If K11 chain formation is inefficient, optimize E2:E3 ratio (test 1:1 to 1:5). For non-specific chains, introduce E2 mutations that favor K11 specificity.
Protocol 3: Computational Optimization of Degraders
Background: Computational approaches enable rational design of protein degraders with optimized binding characteristics and reduced off-target effects [33] [38].
Materials:
- Molecular docking software (Discovery Studio 2019)
- GROMACS 2020.3 for MD simulations
- SwissTargetPrediction database
- Pharmacophore modeling tools
Methodology:
- Target Identification: Input compound structures into SwissTargetPrediction (species: Homo sapiens) to identify potential therapeutic targets.
- Molecular Docking: Perform docking with CHARMM force field to refine ligand shapes and charge distribution. Filter targets with LibDock scores >130 [38].
- Dynamics Simulation: Run 100 ns MD simulations using GROMACS to analyze protein-ligand complex stability. Monitor RMSD, RMSF, and binding free energies (MM-PBSA).
- Pharmacophore Modeling: Construct 3D-QSAR models based on spatial diversity of active compounds. Use these to screen compound libraries with similar features.
- Validation: Synthesize top candidates and evaluate in MCF-7 breast cancer cells, comparing ICâ‚…â‚€ values to positive controls (e.g., 5-FU) [38].
Troubleshooting: If binding poses are unrealistic, adjust flexible residue settings. For poor correlation between docking scores and activity, include solvation effects in binding energy calculations.
Signaling Pathways and Experimental Workflows
Nedd4-2 Autoinhibition and Activation Pathway
Diagram 1: Nedd4-2 Regulation Pathway. This pathway illustrates the transition from autoinhibited to active Nedd4-2 states, highlighting key structural rearrangements triggered by calcium-dependent membrane binding that enable K63-linked polyubiquitination [36].
MARUbylation and K11 Extension Workflow
Diagram 2: MARUbylation Cascade. This workflow details the sequential enzymatic activities in MARUbylation, from initial PARP7 auto-MARylation through DTX2-mediated MARUbe formation to RNF114-dependent K11 polyubiquitin chain extension [35].
Research Reagent Solutions
Table 3: Essential Research Reagents for Ubiquiton Studies
| Reagent/Category | Specific Examples | Function/Application | Considerations |
|---|---|---|---|
| E3 Ligase Tools | Recombinant Nedd4-2, RNF114, ZNF598, DTX2 | Structural studies, biochemical assays, ubiquitination profiling | Confirm phosphorylation status; validate autoinhibition state [36] |
| Linkage-Specific Reagents | K11-linkage specific antibodies, Ub-ADPr fluorescent probes, Ub mutants (K11R, K48R, etc.) | Detection and manipulation of specific ubiquitin chain types | Verify antibody specificity with linkage arrays; optimize probe concentration [35] |
| Computational Resources | AlphaFold3 models, GROMACS, SwissTargetPrediction, Pharmacophore models | Predicting protein interactions, complex stability, and compound screening | Cross-validate computational predictions with experimental data [35] [38] |
| Cell-Based Assay Systems | hiPS cells, MCF-7 breast cancer cells, neuronal differentiation models | Functional validation in relevant biological contexts | Monitor cell-type specific dependencies; assess differentiation state [37] [38] |
| Delivery Technologies | Degrader-Antibody Conjugates (DACs), TfR-binding modules, nanoparticle formulations | Enhancing tissue penetration and cellular uptake | Optimize linker chemistry; assess payload release efficiency [33] |
The strategic optimization of challenging protein targets requires a multidisciplinary approach that integrates structural biology, computational design, and mechanistic enzymology. By leveraging the Ubiquiton inducible linkage-specific polyubiquitylation system alongside detailed understanding of E3 ligase regulation, researchers can overcome historical limitations in targeting recalcitrant proteins. The protocols and analytical frameworks presented herein provide a roadmap for developing next-generation degraders with enhanced specificity and efficacy, ultimately expanding the druggable proteome for therapeutic intervention. Future directions will focus on expanding the E3 ligase toolbox, improving tissue-specific delivery, and refining computational prediction of degradation efficiency to accelerate the translation of these technologies to clinical applications.
The posttranslational modification of proteins by ubiquitin is a fundamental regulatory mechanism that controls a vast array of cellular processes, including protein degradation, signal transduction, DNA repair, and membrane trafficking [15] [39] [7]. The ubiquitin code's complexity arises from the ability of ubiquitin to form polymeric chains through its internal lysine residues (K6, K11, K27, K29, K33, K48, K63) or its N-terminal methionine (M1), with each linkage type generating a functionally distinct signal [39] [40]. For instance, K48-linked chains primarily target substrates for proteasomal degradation, whereas K63-linked chains and M1-linear chains play crucial roles in inflammatory signaling and endocytosis [7] [40]. Mono-ubiquitylation, the attachment of a single ubiquitin moiety, also exerts diverse non-proteolytic functions, particularly in histone regulation and epigenetic control [39].
The Ubiquiton system represents a breakthrough synthetic biology tool that addresses a long-standing experimental challenge: the controlled, inducible polyubiquitylation of proteins of interest with defined linkage specificity in living cells [15] [7] [8]. This innovative system comprises engineered ubiquitin protein ligases paired with matching ubiquitin acceptor tags that enable researchers to precisely induce linear (M1-), K48-, or K63-linked polyubiquitylation on target proteins [7]. The development of such a tool is particularly significant for dissecting the signaling functions of specific ubiquitin chains in diverse biological contexts, from proteasomal targeting to membrane receptor internalization [15]. However, implementing the Ubiquiton system necessitates rigorous validation to confirm that the intended chain linkage is formed and to distinguish between mono- and poly-ubiquitylation events, which this application note addresses in detail.
The Ubiquiton system employs a modular design centered on engineered E3 ubiquitin ligases and specialized substrate tags that work in concert to achieve linkage-specific ubiquitylation. E3 ubiquitin ligases are typically categorized based on their structural features and catalytic mechanisms, primarily as HECT-type, RING-type, or RBR-type ligases [40] [41]. HECT-type ligases, such as the well-characterized HACE1, form a catalytic thioester intermediate with ubiquitin before transferring it to the substrate, while RING-type ligases facilitate direct ubiquitin transfer from E2 enzymes to substrates [42] [40]. The Ubiquiton system likely leverages these distinct catalytic properties to achieve linkage specificity.
The system's functionality relies on several key components that must be thoroughly validated:
- Engineered E3 Ligases: Modified ubiquitin ligases designed to synthesize specific ubiquitin chain linkages (M1, K48, or K63) on target proteins [7].
- Ubiquitin Acceptor Tags: Short peptide sequences fused to proteins of interest that serve as specific substrates for the engineered E3 ligases [8].
- Induction Mechanism: A precisely controlled system (potentially chemical or genetic) that triggers the ubiquitylation reaction only upon activation, enabling temporal analysis [7].
- Cellular Expression System: Platforms for deploying the tool in both yeast and mammalian cells, allowing for cross-organismal validation [7] [8].
The following diagram illustrates the core working principle of the Ubiquiton system and the validation approaches detailed in this application note:
Experimental Protocols for Specificity Validation
Linkage-Specific Antibody Validation
Linkage-specific ubiquitin antibodies provide a rapid and accessible method for initial validation of Ubiquiton system output.
Protocol:
- Sample Preparation:
- Transfert cells with Ubiquiton system components (engineered E3 ligase and tagged protein of interest).
- Induce ubiquitylation according to system specifications (time and concentration optimization may be required).
- Lyse cells in RIPA buffer (or specified lysis buffer) supplemented with 10 mM N-ethylmaleimide (NEM) to preserve ubiquitin conjugates.
- Clarify lysates by centrifugation at 16,000 × g for 15 minutes at 4°C.
Immunoblotting:
- Separate proteins by SDS-PAGE (8-12% gradient gels recommended) and transfer to PVDF membranes.
- Block membranes with 5% non-fat milk in TBST for 1 hour at room temperature.
- Incubate with primary linkage-specific antibodies (see Table 1 for recommendations) diluted in blocking buffer overnight at 4°C.
- Wash membranes 3× with TBST for 10 minutes each.
- Incubate with appropriate HRP-conjugated secondary antibodies for 1 hour at room temperature.
- Develop using enhanced chemiluminescence substrate and image.
Controls:
- Include samples expressing catalytic dead E3 ligase mutants.
- Test Ubiquiton system with non-cognate ubiquitin acceptor tags.
- Express proteins of interest without ubiquitin acceptor tags.
Troubleshooting:
- High background signal may require increased stringency washes (higher salt concentration or addition of 0.1% SDS).
- Weak signal may indicate poor induction efficiency; optimize induction conditions and verify component expression.
Tandem Ubiquitin Binding Entity (TUBE) Pull-Down Assay
TUBE assays enable enrichment of ubiquitylated proteins while protecting against deubiquitylation, providing material for downstream linkage analysis.
Protocol:
- Ubiquitylated Protein Enrichment:
- Prepare cell lysates as described in protocol 3.1.
- Incubate clarified lysates with appropriate TUBE reagents (see Table 2) for 2-4 hours at 4°C with gentle rotation.
- Add appropriate affinity resin (e.g., agarose beads) and incubate for an additional 1-2 hours.
- Pellet beads by gentle centrifugation (500 × g for 3 minutes) and wash 3-4 times with ice-cold lysis buffer.
- Elute bound proteins with 2× SDS-PAGE sample buffer containing 20 mM DTT by heating at 95°C for 10 minutes.
Mass Spectrometry Sample Preparation:
- Separate eluted proteins by short SDS-PAGE (run into gel approximately 1 cm).
- Excise entire gel lanes and process for in-gel digestion.
- Reduce proteins with 10 mM DTT for 30 minutes at 56°C.
- Alkylate with 55 mM iodoacetamide for 30 minutes at room temperature in the dark.
- Digest with trypsin (1:20 enzyme-to-protein ratio) overnight at 37°C.
- Extract peptides with 50% acetonitrile/5% formic acid and dry in a vacuum concentrator.
LC-MS/MS Analysis:
- Reconstitute peptides in 0.1% formic acid.
- Analyze by nanoLC-MS/MS using a long gradient (60-120 minutes) for deep coverage.
- Use data-dependent acquisition methods with dynamic exclusion for comprehensive ubiquitin remnant peptide detection.
Critical Considerations:
- Include control samples with proteasome inhibitor (e.g., MG132) to stabilize ubiquitylated proteins if degradation is observed.
- Use ubiquitin KO cells if available to reduce endogenous ubiquitin background.
Fluorescence-Based Ubiquitylation Assay
Adapted from the recently developed high-sensitivity UBE3A assay [43], this protocol enables rapid, quantitative detection of ubiquitylation activity.
Protocol:
- Assay Setup:
- Express Ubiquiton components with fluorescently tagged ubiquitin or substrates.
- Alternatively, use anti-ubiquitin antibodies with fluorescent secondary antibodies for detection.
- Seed cells in 96-well optical-bottom plates for standardized measurement.
Quantification:
- Induce ubiquitylation and monitor fluorescence intensity over time (0-60 minutes) using a microplate reader.
- Use appropriate filter sets for the fluorescent tags employed.
- Include control wells without induction, without E3 ligase, and with catalytic dead E3 mutants.
Data Analysis:
- Normalize fluorescence values to protein concentration or cell number controls.
- Calculate kinetics parameters (initial velocity, maximum signal) to compare efficiency across different Ubiquiton system variants.
Advantages:
- Enables detection in approximately 1 hour [43].
- Amenable to high-throughput screening of Ubiquiton system variants or small molecule modulators.
- Lower cost compared to mass spectrometry-based methods.
The experimental workflow for a comprehensive specificity validation strategy is visualized below:
Data Analysis and Interpretation
Quantitative Assessment of Ubiquitylation
The validation approaches outlined above generate both qualitative and quantitative data that must be systematically analyzed to confirm Ubiquiton system specificity. Linkage-specific immunoblots provide semi-quantitative data on chain type abundance, while fluorescence-based assays generate precise kinetic measurements of ubiquitylation efficiency. Mass spectrometry delivers the most comprehensive dataset, enabling identification of exact modification sites and relative quantification of different ubiquitin chain linkages.
For mass spectrometry data analysis, specialized bioinformatics tools are essential:
- MaxQuant: Configured to include ubiquitin remnant diGly (K-ε-GG) signature as a variable modification.
- Skyline: For targeted analysis of specific ubiquitin linkage peptides.
- Custom Scripts: To calculate relative abundances of different ubiquitin chain types.
Data normalization is critical for accurate interpretation:
- Normalize ubiquitin signal to total protein input or housekeeping proteins.
- For temporal studies, express data as fold-change relative to time zero.
- Include spike-in standards (e.g., heavy labeled ubiquitin) for precise quantification in mass spectrometry experiments.
Distinguishing Mono- vs. Poly-ubiquitylation
A key validation step for the Ubiquiton system is confirming that it generates polyubiquitin chains rather than monoubiquitylation, as each modification type has distinct functional consequences [39]. Several experimental approaches enable this distinction:
Molecular Weight Analysis:
- Monitor apparent molecular weight shifts by SDS-PAGE: monoubiquitylation typically increases molecular weight by ~8 kDa, while polyubiquitylation creates higher molecular weight smears or discrete bands.
- Use high-percentage gels (10-15%) for better resolution of modified species.
Ubiquitin Laddering Assay:
- Express His-tagged ubiquitin with the Ubiquiton system.
- Purify ubiquitylated proteins under denaturing conditions (e.g., 6 M guanidine hydrochloride).
- Analyze eluates by immunoblotting with anti-ubiquitin antibodies to detect characteristic ubiquitin ladders.
Limited Proteolysis:
- Treat immunoprecipitated ubiquitylated proteins with low concentrations of trypsin.
- Monitor degradation pattern by immunoblotting; polyubiquitylated proteins show a characteristic laddering pattern even after limited proteolysis.
Table 1: Key Characteristics of Mono- vs. Poly-ubiquitylation
| Parameter | Monoubiquitylation | Polyubiquitylation |
|---|---|---|
| Molecular Weight Shift | ~8 kDa increase | Large increases (>16 kDa), often smeared |
| Functional Consequences | Altered activity, localization, interactions | Often degradation (K48) or signaling (K63, M1) |
| Proteasome Sensitivity | Not targeted for degradation | K48-linked chains target to proteasome |
| Antibody Recognition | Single ubiquitin epitope | Multiple ubiquitin epitopes, enhanced signal |
| MS Signature | Single diGly modification on substrate | Multiple diGly modifications, ubiquitin-ubiquitin linkages |
Expected Results and Acceptance Criteria
For the Ubiquiton system to be considered validated, experimental data should meet the following acceptance criteria:
Linkage Specificity:
- >90% of detected ubiquitin chains should match the intended linkage type.
- <5% cross-reactivity with non-cognate linkage types in controlled experiments.
- Consistent linkage specificity across multiple protein substrates.
Induction Response:
- Minimal background ubiquitylation in non-induced controls.
- Dose-dependent increase in ubiquitylation with induction signal.
- Rapid onset of modification (minutes to hours depending on system design).
Functional Validation:
- K48-linked ubiquitylation should decrease substrate half-life in cycloheximide chase assays.
- K63-linked ubiquitylation should alter subcellular localization without promoting degradation.
- M1-linear ubiquitylation should activate NF-κB signaling in appropriate reporter assays.
Research Reagent Solutions
Successful implementation of the Ubiquiton validation protocols requires specific reagents and tools. The following table summarizes essential research reagents with their applications and considerations:
Table 2: Essential Research Reagents for Ubiquiton Validation
| Reagent Category | Specific Examples | Application | Key Features |
|---|---|---|---|
| Linkage-Specific Antibodies | Anti-K48 ubiquitin, Anti-K63 ubiquitin, Anti-linear ubiquitin | Immunoblotting, Immunofluorescence | Validate specific chain linkages; some require special fixation |
| Ubiquitin Affinity Reagents | TUBE (Tandem Ubiquitin Binding Entities), K63-TUBE, diUb probes | Enrichment of ubiquitylated proteins | Protect against DUBs; linkage-specific variants available |
| Mass Spectrometry Reagents | diGly remnant antibody, Heavy labeled ubiquitin standards, Trypsin | Ubiquitin site mapping and quantification | Enable system-wide ubiquitinome analysis |
| Activity Assay Components | Fluorescent ubiquitin substrates, UBE1/E1 enzyme, UbcH5b/E2 enzyme | In vitro ubiquitylation assays | Reconstitute ubiquitylation cascade; measure kinetics |
| Proteasome Inhibitors | MG132, Bortezomib, Carfilzomib | Stabilize ubiquitylated proteins | Prevent degradation of polyubiquitylated substrates |
| Deubiquitylase Inhibitors | PR-619, N-Ethylmaleimide (NEM) | Preserve ubiquitin conjugates | Broad-spectrum or specific DUB inhibitors |
| Positive Control Plasmids | Wild-type ubiquitin, Linkage-specific ubiquitin mutants, Known E3 ligases | Assay validation | Verify antibody specificity and assay performance |
Applications in Drug Discovery and Development
The validated Ubiquiton system holds significant promise for drug discovery and development workflows, particularly in targeted protein degradation and pathway-specific pharmacology. By enabling precise control over protein ubiquitylation, researchers can:
Model Disease-Associated Ubiquitin Signaling:
- Recapitulate pathological ubiquitin signaling events in cellular models.
- Study the functional consequences of specific ubiquitin chain types in disease-relevant contexts.
- Validate putative E3 ligase-substrate relationships implicated in human diseases.
Screen for Ubiquitin Pathway Modulators:
- Develop high-throughput screens for small molecules that enhance or inhibit specific ubiquitylation events.
- Identify compounds that modulate E3 ligase activity toward specific substrates.
- Discover molecular glues that induce neo-substrate ubiquitylation.
Optimize Targeted Protein Degraders:
- Validate PROTAC molecule efficacy and mechanism of action.
- Engineer degron systems with optimized degradation kinetics.
- Study resistance mechanisms to targeted protein degradation therapies.
The ability to precisely control ubiquitin chain linkage formation with the Ubiquiton system, once properly validated using the methods described herein, provides a powerful platform for advancing both basic research and therapeutic development in the ubiquitin-proteasome system field.
Best Practices for Data Interpretation and Experimental Controls
The ubiquitin (Ub) and ubiquitin-like protein (Ubl) conjugation system is a crucial post-translational modification pathway that regulates diverse cellular processes, including protein degradation, signal transduction, and DNA repair [44]. This enzymatic cascade is orchestrated by the sequential action of E1 activating enzymes, E2 conjugating enzymes, and E3 ligases, which work in concert to attach Ub/Ubl molecules to specific substrate proteins [44]. Research in this field, particularly focusing on inducible linkage-specific polyubiquitylation tools, requires sophisticated experimental designs, precise controls, and rigorous data interpretation methods to generate reliable, reproducible findings that can advance both basic science and drug discovery efforts.
Key Experimental Controls for Ubiquitin Research
Fundamental Control Concepts
Experimental controls are intentional sets of experimental conditions that encode scientific assumptions based on prior knowledge, serving to identify or reduce unwanted variation and better isolate intended causal effects [45]. In basic sciences, researchers frequently achieve this using "negative" and "positive" controls, which are additional experimental components designed to detect systematic sources of variation [45]. Proper implementation of controls helps minimize both systematic and random error, leading to more precise estimates and preventing misleading conclusions [45].
Control Applications in Ubiquitin Studies
| Control Type | Definition | Ubiquitin Research Application | Interpretation |
|---|---|---|---|
| Treatment Control | Varies treatment assignment while holding outcome measurement constant [45] | Comparing ubiquitination levels with vs. without proteasome inhibitor MG132 | Confirms observed polyubiquitination leads to proteasomal degradation |
| Outcome Control | Holds treatment constant while varying outcome measurement [45] | Measuring both target protein ubiquitination and unrelated protein phosphorylation | Verifies specificity of ubiquitination effects |
| Contrast Control | Makes assumptions about causal effects to rule out alternative explanations [45] | Using catalytically inactive E3 ligase mutant alongside wild-type | Confirms observed effects require functional ubiquitin ligase activity |
| Negative Control | Conditions where no response is expected [45] | Transfection with empty vector or non-targeting siRNA | Identifies background signals and non-specific effects |
| Positive Control | Conditions where a known response is expected [45] | Treatment with established ubiquitin pathway activator | Verifies experimental system is functioning correctly |
Data Interpretation Framework
Interpretation Challenges in Ubiquitin Research
Data interpretation involves assigning meaning to raw data to produce actionable insights [46]. In ubiquitin research, specific challenges include the transient nature of enzymatic intermediates, the complexity of polyubiquitin chain topologies, and the low steady-state levels of many conjugation intermediates [44]. The Ub/Ubl activation and conjugation processes proceed through formation and rapid dissolution of transient complexes and reaction intermediates that exist at low levels and are often too chemically labile or technically difficult to isolate and examine directly [44].
Systematic Interpretation Approach
A robust framework for interpreting ubiquitination data involves:
Data Sorting and Categorization: Breaking down experimental results into various "data points" or categories for analysis, such as ubiquitination levels, chain linkage types, subcellular localization, and functional consequences [46]. This process creates "buckets" to sort collected data into more meaningful information for interpretation.
Pattern Identification: Systematically examining sorted data for consistent trends, such as time-dependent increases in polyubiquitination, linkage-specific chain formation, or substrate specificity patterns [46]. These patterns form the basis for answering research inquiries.
Connection Analysis: Determining whether observed patterns meaningfully correlate with other cellular processes or experimental manipulations [46]. In ubiquitin research, this might involve connecting specific E3 ligase activities with substrate degradation kinetics or signaling pathway activation.
Essential Methodologies and Protocols
Activity-Based Probing of Ubiquitin Conjugation
Activity-based probes (ABPs) are crucial chemical tools for studying ubiquitin conjugation enzymes, particularly for capturing transient reaction intermediates [44]. These probes typically feature:
Reactive Group ("Warhead"): Forms covalent bonds with enzyme active sites, often employing electrophilic moieties that react with nucleophilic thiols of cysteines in E1, E2, and some E3 active sites [44].
Recognition Element: Typically ubiquitin or ubiquitin-like proteins that confer specific binding to target enzymes [44].
Reporter Group: Facilitates detection and isolation, often including fluorophores or affinity tags [44].
Protocol: ABP Validation for E1-E2-E3 Enzymes
Probe Preparation: Synthesize or obtain Ub/Ubl-based ABPs with C-terminal reactive groups (e.g., vinyl sulfones, acyl phosphates) and appropriate tags (e.g., biotin, FLAG, fluorescent tags) [44].
Lysate Preparation: Generate cell lysates under non-denaturing conditions to preserve enzymatic activities.
Labeling Reaction: Incubate lysates (20-50 μg protein) with ABPs (1-10 μM) in reaction buffer (50 mM Tris-HCl, pH 7.5, 5 mM MgCl₂, 2 mM ATP) for 30-60 minutes at 30°C [44].
Control Reactions: Include parallel reactions with:
- Excess native Ub/Ubl (100-200 μM) to compete ABP labeling
- Catalytically inactive enzyme mutants
- Pre-treatment with specific pathway inhibitors
Detection and Analysis: Resolve proteins by SDS-PAGE, followed by in-gel fluorescence scanning or western blotting with appropriate tag antibodies.
Inducible Linkage-Specific Polyubiquitination Assay
Protocol: Reconstitution of Defined Ubiquitin Chain Formation
Component Purification: Express and purify E1 enzyme, specific E2 conjugating enzyme, E3 ligase of interest, and ubiquitin variants (wild-type and linkage-specific mutants) [44].
Reaction Setup: Combine in 50 μL reaction volume:
- 50 mM Tris-HCl, pH 7.5
- 5 mM MgClâ‚‚
- 2 mM ATP
- 0.1-0.5 μM E1 enzyme
- 1-5 μM E2 conjugating enzyme
- 0.5-2 μM E3 ligase
- 10-50 μM ubiquitin (or defined ubiquitin mutants)
- 1-10 μM substrate protein
Time Course Analysis: Incubate at 30°C and remove aliquots at 0, 5, 15, 30, and 60 minutes.
Reaction Termination: Add SDS-PAGE sample buffer containing 50 mM DTT and heat at 95°C for 5 minutes.
Product Analysis: Resolve by SDS-PAGE and detect by western blotting with anti-ubiquitin and substrate-specific antibodies.
Control Reactions: Essential controls include:
- Minus E1, E2, or E3
- ATP-depleted conditions
- Catalytically dead E2 or E3 mutants
- Ubiquitin mutants defective in specific linkages
Quantitative Data Analysis and Presentation
Key Ubiquitination Assay Parameters
| Parameter | Measurement Method | Typical Range | Interpretation Guidelines |
|---|---|---|---|
| Ubiquitination Efficiency | Western blot densitometry | 5-80% substrate modified | <10%: Low efficiency; >50%: High efficiency |
| Reaction Kinetics | Time-course analysis | Kₘ: 0.1-50 μM ubiquitin | Lower Kₘ indicates tighter enzyme-ubiquitin binding |
| Chain Linkage Specificity | Linkage-specific antibodies or ubiquitin mutants | Varies by E2/E3 combination | Preferred linkage indicates biological function |
| Enzyme Specificity | Multiple substrate testing | 1-100-fold preference | Higher specificity suggests physiological relevance |
| Cellular Turnover | Cycloheximide chase + proteasome inhibition | tâ‚/â‚‚: minutes to hours | Shorter tâ‚/â‚‚ indicates functional degradation |
Statistical Considerations
Robust data interpretation in ubiquitin research requires appropriate statistical approaches:
Replication Strategy: Minimum of three independent biological replicates for each experiment to account for natural variation [47].
Normalization Methods: Use internal controls (e.g., total protein staining, housekeeping proteins) to account for loading variations.
Significance Testing: Apply appropriate statistical tests (t-tests, ANOVA) with multiple comparison corrections where necessary.
Visualization of Ubiquitin Signaling Pathways
Ubiquitin Conjugation Cascade
Experimental Workflow for Inducible Polyubiquitination
Research Reagent Solutions
| Reagent Category | Specific Examples | Function/Application | Key Considerations |
|---|---|---|---|
| Activity-Based Probes | Ub~vinyl sulfone, Ub~acyl phosphate [44] | Trapping active enzyme intermediates | Warhead chemistry affects specificity and reactivity |
| Linkage-Specific Ubiquitin Mutants | K48R, K63R, K48-only, K63-only [44] | Defining chain linkage specificity | Maintain structural integrity while blocking specific lysines |
| E1/E2/E3 Inhibitors | PYR-41 (E1 inhibitor), CC0651 (E2 inhibitor) [44] | Pathway perturbation studies | Verify specificity through counter-screens |
| Tandem Ubiquitin-Binding Entities (TUBEs) | Tandem UBA domains [44] | Enriching polyubiquitinated proteins | Variable affinity for different chain linkages |
| DUB Inhibitors | PR-619 (broad-spectrum), USP-specific inhibitors [44] | Stabilizing ubiquitination events | Consider selectivity and cellular toxicity |
| Reconstitution System Components | Purified E1, E2, E3 enzymes [44] | In vitro ubiquitination assays | Maintain proper folding and catalytic activity |
Troubleshooting and Quality Assurance
Common Experimental Issues
Low Ubiquitination Efficiency: Verify ATP concentration (2 mM optimal), check enzyme activities, confirm proper reaction pH (7.4-7.6), and ensure fresh DTT (1-2 mM) to maintain reducing conditions [44].
High Background Signals: Include appropriate negative controls, optimize antibody concentrations, increase wash stringency, and verify reagent specificity.
Inconsistent Results Between Replicates: Standardize enzyme preparation methods, control reaction temperatures precisely, and use fresh ATP solutions.
Data Quality Assessment
Implement rigorous quality control measures including:
Positive Control Validation: Each experiment should include a well-characterized ubiquitination reaction (e.g., p53 ubiquitination by MDM2) to verify system functionality.
Negative Control Assessment: Monitor background signals in minus-enzyme controls to identify non-specific interactions.
Quantitative Standards: Include reference samples with known ubiquitination levels for calibration where possible.
Implementing robust experimental controls and rigorous data interpretation frameworks is essential for advancing research in the ubiquitin field, particularly for developing and characterizing inducible linkage-specific polyubiquitylation tools. The methodologies and guidelines presented here provide a foundation for generating reliable, reproducible data that can withstand critical scrutiny and contribute meaningfully to both basic science and drug discovery efforts. As the toolset for ubiquitin research continues to expand, maintaining stringent standards for experimental design and data interpretation will remain paramount for translating biochemical findings into biological insights and therapeutic applications.
Benchmarking Ubiquiton: Validation and Comparison to Existing Methods
Within the framework of research on the Ubiquiton inducible linkage-specific polyubiquitylation tool, functional validation in the critical pathways of proteasomal degradation and endocytosis is paramount. These pathways represent two major functional outputs for ubiquitin signaling, controlling protein turnover and cellular trafficking. This application note provides detailed protocols for quantitatively assessing how linkage-specific polyubiquitin chains, generated by the Ubiquiton system, direct substrates toward proteasomal degradation or regulate their endocytic fate. The methodologies outlined herein enable researchers to systematically decipher the ubiquitin code in living cells, with particular emphasis on quantitative mass spectrometry, linkage-specific ubiquitin binding entities, and high-resolution imaging techniques.
Proteasomal Degradation Pathway Analysis
The proteasome primarily recognizes and degrades proteins tagged with specific polyubiquitin chains. While K48-linked chains have been historically characterized as the principal degradation signal, recent quantitative studies reveal that non-K63 linkages collectively mediate proteasomal targeting, with K11-linked chains being particularly abundant and functionally significant [48].
Quantitative Analysis of Polyubiquitin Linkages
Background: Different ubiquitin chain linkages exhibit distinct abundances and respond differently to proteasomal inhibition, indicating their specialized roles in degradation pathways.
Protocol: Isotope Dilution Mass Spectrometry for Linkage Quantification
Cell Lysis and Ubiquitin Conjugate Isolation:
- Lyse cells in denaturing buffer (e.g., 6 M Guanidine-HCl, pH 8.0)
- Enrich ubiquitinated proteins using Ni-NTA affinity chromatography (for His-tagged ubiquitin) or immunoaffinity purification with ubiquitin antibodies [11]
Trypsin Digestion:
- Digest purified ubiquitin conjugates with sequencing-grade trypsin (1:50 enzyme-to-substrate ratio) at 37°C for 16 hours
- This cleaves ubiquitin, leaving a di-glycine (GG) remnant (mass shift of 114.0429 Da) on modified lysine residues [48]
Mass Spectrometry Analysis with Heavy Isotope Standards:
- Spritz samples with chemically synthesized, heavy isotope-labeled, GG-tagged internal standard peptides
- Analyze by LC-MS/MS using multiple reaction monitoring (MRM)
- Quantify native peptides by comparing peak areas with heavy standards [48]
Data Interpretation:
- Calculate absolute amounts of each linkage type using standard curves
- Normalize values to total protein content
Table 1: Average Abundance of Polyubiquitin Linkages in Log-Phase Yeast Cells
| Linkage Type | Percent Abundance (Mean ± SD) |
|---|---|
| K6 | 10.9% ± 1.9% |
| K11 | 28.0% ± 1.4% |
| K27 | 9.0% ± 0.1% |
| K29 | 3.2% ± 0.1% |
| K33 | 3.5% ± 0.1% |
| K48 | 29.1% ± 1.9% |
| K63 | 16.3% ± 0.2% |
Table 2: Fold-Increase in Polyubiquitin Linkages After Proteasomal Inhibition (2-hour treatment)
| Linkage Type | MG132 (100 μM) | PS341 (30 μM) |
|---|---|---|
| K48 | ~8-fold | Similar to MG132 |
| K6, K11, K29 | 4-5-fold | Similar to MG132 |
| K27, K33 | ~2-fold | Similar to MG132 |
| K63 | No significant change | No significant change |
Functional Validation of Proteasomal Targeting
Protocol: Assessing Linkage-Specific Substrate Degradation Using TUBE-Based Assays
Cell Treatment and Lysis:
- Treat cells with proteasome inhibitors (e.g., 100 μM MG132) or vehicle control for 2-4 hours
- Lyse cells in buffer preserving polyubiquitination (e.g., with 1% NP-40, protease inhibitors, and 10 mM N-ethylmaleimide to inhibit DUBs) [13]
Linkage-Specific Ubiquitin Capture:
- Incubate cell lysates with chain-specific Tandem Ubiquitin Binding Entities (TUBEs) immobilized on magnetic beads
- Use Pan-TUBEs, K48-TUBEs, or K63-TUBEs for specific capture
- Incubate for 2 hours at 4°C with gentle rotation [13]
Target Protein Detection:
- Wash beads extensively with lysis buffer
- Elute bound proteins with SDS sample buffer
- Analyze by immunoblotting with target-specific antibodies
Application Example:
- For RIPK2 analysis: Stimulate THP-1 cells with L18-MDP (200 ng/mL, 30 min) for K63-ubiquitination or treat with RIPK2 PROTAC (e.g., RIPK degrader-2) for K48-ubiquitination [13]
- Capture with linkage-specific TUBEs and detect with anti-RIPK2 antibody
Diagram 1: Ubiquiton-induced polyubiquitin chains direct substrates to the proteasome. K48, K11, and other non-K63 linkages function as proteasomal targeting signals.
Endocytosis and Trafficking Pathway Analysis
Ubiquitin serves as a critical signal for membrane protein internalization and endosomal sorting, typically through monoubiquitination or K63-linked chains, though other linkages may also participate [49].
Monitoring Ubiquitin-Dependent Endocytosis
Background: Ubiquitin modification of membrane proteins recruits adaptor proteins that facilitate clathrin-mediated endocytosis and subsequent sorting to multivesicular bodies (MVBs) via the ESCRT machinery.
Protocol: Live-Cell Imaging of Endocytic Protein Dynamics
Sample Preparation:
- Engineer cells to express GFP-tagged endocytic proteins (e.g., clathrin, Sla2, Abp1) from endogenous genomic loci
- Co-express ubiquitin or Ubiquiton tools with specific linkage capabilities [50]
Image Acquisition:
- Use wide-field epifluorescence microscopy with high-sensitivity cameras
- Acquire videos at the equatorial plane of cells
- Maintain temperature at appropriate level (e.g., 30°C for yeast) throughout imaging [50]
Data Analysis:
- Track centroid positions of fluorescent patches over time
- Align individual trajectories spatially and temporally using specialized algorithms
- Calculate average trajectories with intensity profiles
- Estimate molecule numbers using kinetochore protein fluorescence for calibration [50]
Key Measurements:
- Protein recruitment timing and sequence
- Movement speed and directionality
- Patch assembly and disassembly kinetics
- Correlation with ubiquitin signals
Ubiquitin-Dependent Receptor Trafficking Assay
Protocol: Tracking Notch Ligand Endocytosis and Signaling
Genetic Manipulation:
- Express wild-type or ubiquitination-deficient mutants of Notch ligands (Delta, Serrate)
- Modulate E3 ligase activity (Neuralized, Mind bomb) or express inhibitory Brd family peptides [49]
Endocytosis Measurement:
- Monitor ligand internalization using antibody uptake assays
- Fix cells at timed intervals after surface labeling
- Process for immunofluorescence and confocal microscopy
Functional Assessment:
- Quantify Notch signaling activity using luciferase reporter assays
- Measure downstream target gene expression (e.g., by qRT-PCR)
- Correlate ligand ubiquitination status with signaling output [49]
Diagram 2: Ubiquitin-dependent endocytosis and trafficking pathway. Ubiquiton-induced ubiquitination directs receptor internalization and lysosomal degradation via the ESCRT machinery.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents for Ubiquitin Pathway Analysis
| Reagent/Tool | Function/Application | Examples/Specifications |
|---|---|---|
| Linkage-Specific TUBEs | High-affinity capture of specific polyubiquitin chains | K48-TUBEs, K63-TUBEs, Pan-TUBEs; nm affinity range [13] |
| Ubiquitin Binding Domains (UBDs) | Enrichment of ubiquitinated proteins | Tandem UBDs for increased affinity; used in ubiquitin proteomics [11] |
| Linkage-Specific Antibodies | Immunodetection of specific ubiquitin linkages | K48-specific, K63-specific, M1-linear specific antibodies [11] |
| Proteasome Inhibitors | Block proteasomal activity to study ubiquitin chain accumulation | MG132 (10-100 μM), PS341 (30 μM) [48] |
| Deubiquitinase (DUB) Inhibitors | Preserve ubiquitin signals during lysis | N-ethylmaleimide (10 mM), PR-619 [13] |
| Epitope-Tagged Ubiquitin | Affinity purification of ubiquitinated proteins | His-tagged Ub, Strep-tagged Ub, HA-Ub [11] |
| Stable Isotope Labels | Quantitative mass spectrometry | SILAC labels, heavy peptide standards for absolute quantification [48] |
Advanced Applications: Studying Branched Ubiquitin Chains
Emerging evidence indicates that branched ubiquitin chains represent a sophisticated layer of regulation in ubiquitin signaling, with unique functions that are not merely the sum of their constituent linkages [51] [52].
Analyzing Branched Chain Functions
Protocol: UbiREAD (Ubiquitinated Reporter Evaluation After Intracellular Delivery)
Substrate Preparation:
- Generate defined ubiquitin chain architectures on model substrates (e.g., GFP) using recombinant enzymes
- Create homotypic chains (K48, K63) and branched chains (K48/K63) of defined length [52]
Intracellular Delivery:
- Deliver bespoke ubiquitinated proteins into human cells using nanoparticle-stabilized nanocapsules or other delivery methods
- Optimize delivery efficiency for each cell type [52]
High-Temporal Resolution Monitoring:
- Monitor substrate degradation and deubiquitination kinetics using fluorescent reporters
- Sample at minute-level intervals to capture rapid early events
- Compare half-lives of differently ubiquitinated substrates [52]
Key Finding Application:
- For K48/K63-branched chains, determine that the substrate-anchored chain identity dictates degradation behavior, establishing a functional hierarchy within branched ubiquitin chains [52]
The protocols detailed in this application note provide a comprehensive framework for functionally validating Ubiquiton tools in the key pathways of proteasomal degradation and endocytosis. By employing quantitative mass spectrometry, linkage-specific affinity tools, and dynamic imaging approaches, researchers can systematically decipher how specific ubiquitin linkages direct cellular outcomes. These methodologies enable the precise characterization of Ubiquiton-induced polyubiquitylation, advancing both fundamental understanding of ubiquitin signaling and the development of targeted degradation therapies.
The controlled degradation of specific proteins is an indispensable tool for understanding protein function and developing new therapeutic strategies. Among the various technologies developed, the Ubiquiton system has emerged as a uniquely precise tool for inducing linkage-specific polyubiquitylation. This application note provides a comparative analysis of Ubiquiton against established degron technologies such as PROTACs, AID, and dTAG, framing the discussion within the broader context of ubiquitin code research. We detail specific experimental protocols and provide a reagent toolkit to facilitate the adoption of these powerful methods by researchers and drug development professionals.
Targeted protein degradation systems harness the cell's own proteolytic machinery but differ fundamentally in their mechanisms, components, and applications.
Core Mechanism Classification
- Ubiquiton System: An engineered, inducible ubiquitin protein ligase system that directly catalyzes the formation of a specific polyubiquitin chain topology (M1-, K48-, or K63-linked) on a target protein fused to a matching ubiquitin acceptor tag [7] [15].
- PROTACs: Heterobifunctional small molecules that bring a target protein into proximity with an E3 ubiquitin ligase, leading to its polyubiquitylation and degradation. Their action depends on endogenous E2 conjugating enzymes and is typically linkage-agnostic [53] [54].
- Classical Degron Systems (AID, dTAG): Relies on fusing a target protein to a degron tag (e.g., AID, FKBP12F36V). The addition of a small molecule (e.g., auxin, dTAG ligand) induces recruitment to a specific E3 ligase complex (e.g., OsTIR1, CRBN), leading to ubiquitylation and degradation [55] [56].
The following diagram illustrates the fundamental mechanistic differences between these systems.
Quantitative Performance Comparison
The table below summarizes the key characteristics of each technology, highlighting their strategic differences.
| Feature | Ubiquiton System | PROTACs | AID 2.0/2.1 | dTAG |
|---|---|---|---|---|
| Core Component | Engineered E3 & acceptor tag [7] | Heterobifunctional small molecule [53] | AID tag + OsTIR1(F74G/S210A) adapter [55] | FKBP12F36V tag + dTAG ligand [55] |
| Degradation Mechanism | Linkage-specific polyubiquitylation [7] | Proximity-induced, linkage-agnostic ubiquitylation [53] [54] | OsTIR1-AID recognition & ubiquitylation [55] | CRBN- or VHL-mediated ubiquitylation [55] |
| Induction | Genetically inducible (e.g., chemically) | Addition of small molecule [54] | Addition of auxin (e.g., 5-Ph-IAA) [55] | Addition of dTAG molecule (e.g., dTAG-13) [55] |
| Kinetics (Degradation) | Rapid, tunable by induction level | Varies by PROTAC & target; can be rapid [54] | Very fast (superior kinetics) [55] | Fast [55] |
| Key Advantage | Precise control over ubiquitin chain linkage; inducible [7] | Targets endogenous proteins; broad potential therapeutic use [53] [54] | High efficiency & reversibility; improved variants reduce basal degradation [55] | High degradation efficiency & reversibility [55] |
| Primary Limitation | Requires genetic fusion of acceptor tag | Limited by E3 ligase availability & off-targets [57] [58] | Requires genetic fusion of AID tag & OsTIR1 expression; some basal degradation [55] | Requires genetic fusion of dTAG; ligand can impact cell viability [55] |
| Therapeutic Applicability | Primarily a research tool | High (Multiple candidates in clinical trials) [54] [59] | Research tool | Research tool |
Experimental Protocols
This section provides detailed methodologies for applying these technologies in a research setting.
Protocol: Applying the Ubiquiton System to Study Proteasomal Targeting
Objective: To induce K48-linked polyubiquitylation of a nuclear protein of interest (POI) and monitor its subsequent degradation by the proteasome.
Background: K48-linked ubiquitin chains are the canonical signal for proteasomal degradation [60]. This protocol uses the Ubiquiton system to directly append this specific degradation signal to a target protein, bypassing the need for endogenous E2/E3 recognition [7].
Materials:
- Plasmids: Ubiquiton K48-specific E3 ligase, POI fused to the matching ubiquitin acceptor tag.
- Cell Line: HEK293T or other suitable mammalian cell line.
- Transfection reagent (e.g., polyethyleneimine (PEI)).
- Inducer (e.g., Doxycycline, if using a tetracycline-inducible promoter).
- Lysis Buffer: RIPA buffer supplemented with protease inhibitors (e.g., Complete Mini EDTA-free) and 10mM N-Ethylmaleimide (NEM) to inhibit deubiquitinases.
- Antibodies: Anti-POI antibody, Anti-Ubiquitin (linkage-specific, e.g., K48), Anti-Tubulin (loading control).
Procedure:
- Cell Seeding and Transfection: Seed HEK293T cells in a 6-well plate. At 60-70% confluency, co-transfect cells with the plasmids encoding the K48-specific Ubiquiton E3 ligase and the POI-acceptor tag fusion using your preferred transfection method.
- Induction of Ubiquitylation: 24 hours post-transfection, induce the expression of the Ubiquiton E3 ligase by adding Doxycycline (e.g., 1 µg/mL) to the culture medium.
- Time-Course Sampling: Harvest cells at various time points post-induction (e.g., 0, 2, 4, 8, 12 hours) by washing with PBS and lysing in ice-cold RIPA buffer with inhibitors.
- Analysis:
- Western Blotting: Resolve proteins by SDS-PAGE and transfer to a PVDF membrane. Probe with anti-POI antibody to monitor the loss of the protein over time. A smear or upward shift indicates ubiquitylation. Re-probe the membrane with anti-K48-linkage specific ubiquitin antibody to confirm the formation of K48-linked chains. Use anti-Tubulin as a loading control.
- Cycloheximide Chase (Optional): To specifically measure protein half-life, treat cells with the protein synthesis inhibitor cycloheximide (e.g., 100 µg/mL) at the time of induction and proceed with the time-course sampling and western blot analysis.
Protocol: Comparing Degradation Kinetics Across Systems
Objective: To quantitatively compare the degradation and recovery kinetics of the AID 2.1 system against dTAG and PROTACs.
Background: A systematic comparison of inducible degron technologies in human iPSCs revealed critical differences in degradation kinetics, basal degradation, and recovery rates, which are crucial for experimental design [55].
Materials:
- Cell Lines: Isogenic hiPSC clonal lines with homozygous C-terminal tags:
- Gene X-AID2.1 + OsTIR1(S210A) at AAVS1 locus.
- Gene X-FKBP12F36V (for dTAG).
- (For PROTAC: Wild-type cells expressing the endogenous target).
- Ligands: 5-Ph-IAA (for AID), dTAG-13 (for dTAG), appropriate PROTAC (e.g., ARV-110 for AR).
- Antibodies: High-specificity antibody against Protein X.
Procedure:
- Degradation Kinetics Assay:
- Seed each cell line in multiple wells of a 12-well plate.
- Once cells reach 70-80% confluency, add the respective ligands: 500 nM 5-Ph-IAA (AID), 1 µM dTAG-13 (dTAG), or a validated concentration of the PROTAC.
- Harvest cells at T = 0, 0.5, 1, 2, 4, 6, and 24 hours post-ligand addition.
- Perform western blot analysis on lysates to quantify Protein X levels. Normalize to a loading control.
- Reversibility/Recovery Assay:
- Treat cells with respective ligands for 6 hours to achieve robust degradation.
- Wash cells thoroughly 3x with pre-warmed PBS to remove the ligand.
- Add fresh ligand-free medium and harvest cells at T = 0 (washout), 6, 12, 24, and 48 hours after washout.
- Analyze Protein X recovery by western blot.
The Scientist's Toolkit: Essential Research Reagents
The following table lists key reagents essential for research in targeted protein degradation.
| Reagent Category | Specific Example | Function & Application |
|---|---|---|
| E3 Ligase Ligands | VHL Ligand (VH-032), CRBN Ligand (Lenalidomide), IAP Ligand (LCL161) | Component of PROTAC molecules for recruiting specific E3 ligases [54]. |
| POI Ligands | AR antagonist (e.g., Enzalutamide), ER antagonist (e.g., Fulvestrant), BET bromodomain inhibitor (e.g., JQ1) | Component of PROTAC molecules for binding the target protein [54]. |
| Degron System Ligands | Auxin (IAA) / 5-Ph-IAA, dTAG-13 / dTAG-7 | Small molecule inducers for AID and dTAG systems, respectively [55]. |
| Linkage-Specific Ubiquitin Antibodies | Anti-K48-Ubiquitin, Anti-K63-Ubiquitin, Anti-M1 (Linear)-Ubiquitin | Critical tools for validating the specific ubiquitin chain topology formed by tools like Ubiquiton [60] [7]. |
| PROTAC Clinical Candidates | ARV-471 (Vepdegestrant), ARV-110, KT-474 | Benchmark molecules for studying PROTAC efficacy, mechanisms, and off-target effects in relevant disease models [54] [59]. |
| Engineered Cell Lines | AID-ready lines (e.g., expressing OsTIR1(F74G/S210A)), dTAG-ready lines | Pre-engineered cellular models that simplify the study of endogenous proteins tagged with specific degrons [55]. |
The choice between Ubiquiton, PROTACs, and classical degrons is dictated by the specific research or therapeutic question.
- Use the Ubiquiton system when the biological question hinges on understanding the functional consequence of a specific ubiquitin chain linkage. Its ability to precisely control chain type makes it unparalleled for deciphering the ubiquitin code [7].
- Employ PROTACs for therapeutic exploration or when targeting endogenous proteins without genetic modification is required. Their pharmacology and the rapidly expanding clinical pipeline (e.g., ARV-471, BMS-986365) make them the leading modality for drug development [54] [59].
- Select classical degron systems (AID, dTAG) for high-speed, reversible protein depletion in fundamental research, especially when using endogenously tagged proteins in genetically tractable model systems. The high efficiency and reversibility of modern AID variants (e.g., AID 2.1) are ideal for studying dynamic biological processes and essential genes [55].
In conclusion, Ubiquiton represents a specialized, powerful tool for mechanistic studies of ubiquitin signaling, complementing rather than replacing the broader applications of PROTACs and other degron technologies. As the field advances, the integration of these tools—using Ubiquiton to define signaling mechanisms and PROTACs to translate these insights into therapies—will powerfully accelerate both basic discovery and drug development.
The Ubiquiton system represents a groundbreaking synthetic biology tool that addresses a fundamental limitation in ubiquitin research: the inability to induce specific polyubiquitin linkages on proteins of interest in living cells. As a posttranslational modification, ubiquitin regulates virtually all cellular processes through the formation of polymeric chains with distinct linkages that determine diverse functional outcomes. Conventional genetic knockdown approaches suffer from compensatory mechanisms, pleiotropic effects, and temporal limitations, whereas Ubiquiton provides precise spatiotemporal control over linkage-specific polyubiquitylation. This system enables researchers to directly interrogate the consequences of M1- (linear), K48-, and K63-linked polyubiquitin chains on target proteins, offering unprecedented specificity for dissecting ubiquitin-dependent signaling pathways in both yeast and mammalian cells [7] [8].
The technological innovation centers on a set of engineered ubiquitin protein ligases and matching ubiquitin acceptor tags that work in concert to achieve rapid, inducible polyubiquitylation. By applying this system to multiple biological contexts including proteasomal targeting and endocytic pathways, researchers have validated its utility for diverse protein types including soluble cytoplasmic, nuclear, chromatin-associated, and integral membrane proteins [7]. The ability to control protein localization and stability with chain-type specificity positions Ubiquiton as a transformative tool for exploring ubiquitin signaling in disease contexts and therapeutic development.
Technical Architecture and Molecular Components
System Design and Core Mechanism
The Ubiquiton system employs a modular architecture consisting of two primary components: engineered E3 ubiquitin ligases and specialized ubiquitin acceptor tags. The E3 ligases are optimized for generating specific ubiquitin linkages: E3(1) for linear (M1-linked), E3(48) for K48-linked, and E3(63) for K63-linked polyubiquitin chains [7]. These engineered ligases overcome the natural promiscuity of endogenous E3 enzymes that typically produce mixed chain types, thereby enabling precise ubiquitin code writing.
The matching ubiquitin acceptor tags (NUbo for N-terminal, CUbo for C-terminal fusion) serve as substrates for the engineered E3 ligases and are fused to proteins of interest. These tags contain optimized recognition motifs that ensure efficient and specific ubiquitylation by their cognate E3 ligases. The system is designed with inducible expression elements (doxycycline, copper) that provide temporal control over the polyubiquitylation process, allowing researchers to initiate chain formation at precise experimental timepoints [61]. This inducibility is crucial for studying dynamic processes like protein degradation and trafficking without the compensatory adaptations that often complicate traditional genetic knockdown approaches.
Research Reagent Solutions
Table 1: Essential Research Reagents for Ubiquiton System Implementation
| Reagent Category | Specific Examples | Function and Application |
|---|---|---|
| Engineered E3 Ligases | myc-E3(1)-CUbo, NUbo-E3(48), NUbo-E3(63) | Induce specific polyubiquitin chain linkages (M1, K48, K63) on target proteins [61] |
| Ubiquitin Acceptor Tags | NUbo-VSV, CUbo-VSV, NUbo-mCherry-VSV | Fused to proteins of interest to serve as substrates for specific polyubiquitylation [61] |
| Detection Antibodies | ANTI-FLAG M2, Anti-GFP, Anti-VSV Glycoprotein, Anti-Ubiquitin (clone FK2) | Detect tagged proteins and ubiquitin modifications [62] |
| Specialized Linkage Antibodies | Anti-ubiquitin Lys63-specific (clone Apu3), Anti-linear ubiquitin (clone LUB9) | Verify specific ubiquitin linkage types in experimental validation [62] |
| Proteasome Inhibitors | Cycloheximide, Chloroquine, Protease inhibitor cocktails | Block protein degradation to assess ubiquitin-mediated turnover [62] |
| Expression Plasmids | pET series (bacterial), YIp series (yeast), inducible systems (TET, CUP1) | Enable system expression in different model organisms [61] |
Experimental Protocols and Applications
Protocol: Inducible Protein Degradation via K48-Linked Ubiquitylation
Purpose: To demonstrate controlled protein degradation through Ubiquiton-mediated K48-linked polyubiquitylation, which targets proteins to the proteasome [7].
Materials:
- Plasmids for NUbo/E3(48) expression (e.g., YIp128-TET-NUbo-E3(48)-VSV) [61]
- Target protein of interest fused to CUbo tag
- Doxycycline for induction
- Cycloheximide to block new protein synthesis [62]
- Anti-VSV antibody for immunoblotting [62]
- K48-linkage specific ubiquitin antibody (optional)
Methodology:
- Cell Preparation: Transfect mammalian cells or generate stable yeast strains expressing both the NUbo-E3(48) ligase and your protein of interest fused to the CUbo acceptor tag [61].
- Induction: Add doxycycline (0.1-1 μg/mL for mammalian cells) to induce expression of the E3(48) ligase component. For yeast systems with copper-inducible promoters, add CuSO₄ (50-100 μM) [61].
- Time Course Sampling: Collect samples at 0, 15, 30, 60, 120, and 240 minutes post-induction.
- Inhibition Control: Treat parallel samples with cycloheximide (100 μg/mL) 10 minutes before induction to block new protein synthesis and isolate degradation effects [62].
- Analysis: Process samples for SDS-PAGE and immunoblotting using VSV antibody to monitor target protein levels. Probe with K48-linkage specific ubiquitin antibody to verify chain type [62].
Expected Results: Rapid depletion of the target protein within 60-120 minutes post-induction, demonstrating efficient proteasomal targeting. Control experiments with catalytic mutant E3(48) (aa24-150) should show stable protein levels [61].
Protocol: Membrane Protein Internalization via K63-Linked Ubiquitylation
Purpose: To control endocytic trafficking of membrane proteins through Ubiquiton-mediated K63-linked polyubiquitylation, mimicking physiological sorting signals [7].
Materials:
- Plasmids for NUbo/E3(63) expression (e.g., YIp128-TET-NUbo-E3(63)-VSV) [61]
- Membrane protein of interest (e.g., EGFR) fused to CUbo tag
- Anti-linear ubiquitin antibody (clone LUB9) [62]
- Fluorescence microscopy system for live-cell imaging
- Temperature-controlled incubation system
Methodology:
- System Setup: Co-express NUbo-E3(63) ligase and membrane protein-CUbo fusion in appropriate cell type.
- Synchronization: Serum-starve cells overnight to minimize background signaling.
- Induction and Imaging: Induce E3(63) expression with doxycycline while performing live-cell imaging at 37°C with 5% CO₂.
- Fixed-Time Analysis: For endpoint measurements, fix cells at 0, 10, 20, and 30 minutes post-induction and process for immunofluorescence.
- Co-localization: Stain with early endosome marker (EEA1) and ubiquitin antibodies to confirm internalization and ubiquitin chain formation [62].
- Control: Include cells expressing catalytically impaired E3(63) (I227A) to demonstrate specificity [61].
Expected Results: Rapid internalization of the target membrane protein within 15-30 minutes, with co-localization in EEA1-positive endosomal compartments. The catalytically impaired E3 should show minimal internalization.
Quantitative Assessment of Ubiquiton Performance
Table 2: Quantitative Performance Metrics of Ubiquiton System Applications
| Experimental Application | Target Protein Type | Induction Time | Efficiency Metric | System Validation |
|---|---|---|---|---|
| Proteasomal Degradation | Soluble cytoplasmic and nuclear proteins | 15-60 minutes | ~70-90% target depletion within 2 hours [7] | Catalytic mutant controls show stable protein levels [61] |
| Endocytic Trafficking | Integral membrane proteins (e.g., EGFR) | 10-30 minutes | ~60-80% internalization within 30 minutes [7] | Co-localization with endosomal markers; linkage-specific antibodies [62] |
| Chromatin Modification | Chromatin-associated proteins | 30-120 minutes | Altered mobility and protein interactions [7] | Chromatin fractionation and co-immunoprecipitation |
| Pathway Activation | Signaling complex components | 5-15 minutes | NF-κB and kinase pathway activation [63] | Reporter assays and phosphorylation status |
Advantages Over Conventional Genetic Knockdown
Linkage Specificity and Signaling Precision
The Ubiquiton system provides unprecedented specificity for studying ubiquitin signaling, overcoming fundamental limitations of conventional genetic approaches. While traditional E3 ligase knockdown or knockout strategies disrupt multiple substrates and signaling pathways, Ubiquiton enables precise formation of defined ubiquitin chain types (M1, K48, K63) on specific target proteins [7]. This specificity is crucial because different ubiquitin linkages encode distinct cellular functions: K48 chains primarily target proteins for proteasomal degradation, K63 chains regulate DNA repair and endocytic trafficking, while linear M1 chains modulate NF-κB signaling and immune responses [7] [63].
The molecular architecture of Ubiquiton ensures this linkage specificity through engineered E3 ligases with defined linkage preferences. For example, the E3(48) component specifically generates K48-linked chains, while E3(63) produces K63-linked chains, and E3(1) creates linear M1-linked ubiquitin polymers [7]. This precision enables researchers to dissect the functional consequences of specific ubiquitin codes without the confounding effects of mixed chain types that occur with endogenous ubiquitylation. Validation experiments using linkage-specific ubiquitin antibodies (e.g., Apu3 for K63-chains, LUB9 for linear chains) confirm the selectivity of the system [62].
Temporal Control and Experimental Flexibility
Unlike constitutive genetic manipulations, Ubiquiton provides precise temporal control through inducible expression systems (doxycycline, copper) that initiate polyubiquitylation at defined timepoints [61]. This temporal resolution enables researchers to study dynamic processes like protein turnover, pathway activation, and trafficking kinetics with minute-scale precision. The system achieves detectable ubiquitin chain formation within 5-15 minutes of induction, with functional outcomes (degradation, internalization) typically observed within 15-60 minutes [7].
The experimental flexibility of Ubiquiton extends to multiple model systems, with optimized vectors for mammalian cells, yeast, and bacterial expression [61]. This cross-platform compatibility facilitates validation in different biological contexts and enables researchers to choose the most appropriate system for their specific questions. The modular design also allows tagging of diverse protein types, including soluble nuclear/cytoplasmic proteins, chromatin-associated factors, and integral membrane proteins, demonstrating broad applicability across different cellular compartments and functional classes [7].
Functional Compensation Avoidance
Conventional genetic knockdown approaches often trigger compensatory mechanisms and adaptive responses that complicate data interpretation. The rapid inducible nature of the Ubiquiton system circumvents these compensatory pathways by creating an acute perturbation that can be measured before cellular adaptation occurs. This is particularly valuable for studying essential genes and pathways where chronic depletion would be lethal or trigger alternative signaling routes.
The system also avoids pleiotropic effects associated with traditional E3 ligase manipulation. Since endogenous E3 enzymes typically modify multiple substrates, their genetic disruption affects numerous downstream pathways. In contrast, Ubiquiton targets specific proteins of interest without globally altering the ubiquitin proteome, providing cleaner experimental readouts and more straightforward interpretation of cause-effect relationships in ubiquitin-dependent processes [7].
Troubleshooting and Technical Considerations
Optimization Guidelines for Challenging Targets
While Ubiquiton demonstrates broad efficacy across diverse protein classes, some targets may require optimization for optimal performance. For refractory substrates, consider testing both N-terminal (NUbo) and C-terminal (CUbo) tagging configurations, as accessibility to the E3 ligase can vary with fusion orientation [61]. For proteins exhibiting incomplete ubiquitylation, increase the expression ratio of E3 ligase to target protein or extend induction times while monitoring for potential overexpression artifacts.
When working with membrane proteins, verify proper localization following tag fusion, as some tags may interfere with trafficking or function. Employ controlled experimentation with catalytic mutant E3 ligases (e.g., E3(1) C885A, E3(63) I227A) to distinguish Ubiquiton-specific effects from background phenotypes [61]. For kinetic studies, include cycloheximide chase experiments to isolate degradation rates from ongoing protein synthesis [62].
Validation and Control Experiments
Rigorous validation is essential for interpreting Ubiquiton experiments correctly. The following control experiments are recommended:
- Catalytic Mutant Controls: Always include parallel experiments with catalytically impaired E3 ligases (e.g., E3(1) C885A, E3(63) I227A) to demonstrate that observed phenotypes require enzymatic activity [61].
- Linkage Specificity Verification: Use linkage-specific ubiquitin antibodies (Apu3 for K63, LUB9 for linear chains) to confirm the intended chain type formation [62].
- Time Course Analyses: Establish comprehensive kinetics for each novel target to identify optimal experimental timepoints.
- Pathway-Specific Inhibitors: Include pharmacological inhibitors (e.g., proteasome inhibitors for degradation studies) to verify the expected mechanistic pathways.
Documentation of expression levels for both E3 components and target proteins is crucial, as excessive expression can lead to non-physiological effects. Quantitative immunoblotting with reference standards enables more accurate comparisons across experiments and conditions.
The Ubiquiton system represents a breakthrough in synthetic biology, fulfilling a long-standing gap in the ubiquitin research field: the ability to induce linkage-specific polyubiquitylation of a protein of interest on demand [64]. This innovative tool combines engineered E3 ubiquitin ligases with a rapamycin-inducible dimerization system and split-ubiquitin technology to achieve precise control over polyubiquitin chain formation [64] [8]. Unlike traditional degradation domain (degron) technologies or PROTACs that often produce heterogeneous ubiquitin chains, Ubiquiton enables researchers to install defined ubiquitin linkages—specifically M1-(linear), K48-, or K63-linked chains—onto target proteins, allowing precise dissection of ubiquitin-dependent signaling pathways [64]. This application note details the implementation, validation, and versatility of the Ubiquiton system across diverse model systems and protein classes, providing researchers with comprehensive protocols to investigate the ubiquitin code in physiological contexts.
Core Mechanism and Components
The Ubiquiton system addresses three fundamental challenges in linkage-specific ubiquitylation: linkage selectivity of conjugation enzymes, inducible substrate selection, and controlled chain initiation [64]. The system employs engineered E3 ligases derived from well-characterized domains with defined linkage specificities: the M1-specific human HOIP, K48-specific yeast Cue1-Ubc7 complex, and K63-specific yeast Pib1-Ubc13·Mms2 complex [64]. These engineered "extender E3s" exhibit exquisite linkage specificity but cannot initiate chains de novo.
To overcome this limitation, Ubiquiton incorporates a split-ubiquitin technology where the N-terminal (NUb, aa 1-37) and C-terminal (CUb, aa 35-76) halves of ubiquitin are brought together via a rapamycin-inducible FKBP-FRB dimerization system [64] [8]. The CUb half contains the specific lysine residue (K48 or K63) that serves as the acceptor site for chain extension, while a mutation (G76V) renders it resistant to cleavage by cellular deubiquitinases (DUBs) [64]. A key refinement was the introduction of an I13A mutation in the NUb module (creating "NUa") to reduce background affinity between the ubiquitin halves, ensuring strict rapamycin control [64].
Table: Core Components of the Ubiquiton System
| Component | Description | Function |
|---|---|---|
| Engineered E3 Ligases | M1-specific (HOIP), K48-specific (Cue1-Ubc7), K63-specific (Pib1-Ubc13·Mms2) | Catalyze linkage-specific polyubiquitin chain extension |
| NUbo Tag | NUa-HA-FRB fusion | Provides N-terminal ubiquitin half and rapamycin-binding domain |
| CUbo Tag | FKBP-CUb(G76V) fusion | Provides C-terminal ubiquitin half with acceptor lysine and rapamycin-binding domain |
| Rapamycin | Small molecule dimerizer | Induces complex assembly by bridging FKBP and FRB domains |
System Architecture and Workflow
The following diagram illustrates the core mechanism of the Ubiquiton system and the experimental workflow for studying its effects:
Applications Across Biological Processes
The Ubiquiton system has been rigorously validated for controlling diverse biological processes through targeted, linkage-specific ubiquitylation. The table below summarizes key applications and their functional outcomes.
Table: Ubiquiton Applications and Functional Outcomes
| Biological Process | Ubiquitin Linkage | Target Protein Class | Functional Outcome | Experimental System |
|---|---|---|---|---|
| Proteasomal Degradation | K48 | Soluble cytoplasmic and nuclear proteins | Rapid protein depletion (t½ ~5-30 min) [64] | Yeast and mammalian cells |
| Endocytic Trafficking | K63 | Plasma membrane receptors (e.g., EGFR) | Ligand-independent internalization [64] | Mammalian cells |
| DNA Damage Tolerance | K63 | Chromatin-associated PCNA | Altered DNA repair dynamics [64] | Budding yeast |
| Inflammatory Signaling | M1/Linear | Signaling complexes | Pathway modulation [64] | Mammalian cells |
Targeted Protein Degradation via K48-Linked Ubiquitylation
Background: K48-linked polyubiquitin chains represent the canonical signal for proteasomal degradation [64] [65]. The Ubiquiton system enables precise control over protein half-life without relying on endogenous E3 ligases that may produce heterogeneous chain types.
Protocol: Inducible Protein Degradation in Mammalian Cells
Molecular Cloning:
- Clone your protein of interest (POI) in-frame with the CUbo tag (FKBP-CUb(G76V)) at either N- or C-terminus
- Co-clone the NUbo (NUa-HA-FRB) fused to the K48-specific E3 ligase (Cue1-Ubc7)
- Include fluorescent tags (e.g., GFP) for live-cell monitoring
Cell Culture and Transfection:
- Culture HEK293T or other appropriate cell lines in standard conditions
- Transfect using preferred method (PEI, calcium phosphate, or lipofection)
- Allow 24-48 hours for protein expression
Induction and Degradation:
- Prepare fresh rapamycin stock solution (1 mM in DMSO)
- Treat cells with 100-500 nM rapamycin or vehicle control
- Monitor degradation kinetics by collecting samples at 0, 5, 15, 30, 60, and 120 minutes post-induction
Analysis:
- Process samples for Western blotting using anti-GFP and loading control antibodies
- Quantify band intensity and calculate half-life using degradation kinetics
- For live-cell imaging, monitor fluorescence intensity over time using time-lapse microscopy
Expected Results: The K48-Ubiquiton system typically achieves rapid protein depletion with half-lives ranging from 5-30 minutes in mammalian cells, depending on the target protein and expression level [64]. This represents a significant improvement over traditional degron systems, with the added benefit of linkage specificity.
Control of Endocytic Trafficking via K63-Linked Ubiquitylation
Background: K63-linked ubiquitin chains primarily function in non-proteolytic signaling, including intracellular trafficking, inflammation, and DNA repair [64] [66]. The Ubiquiton system enables direct testing of whether K63 ubiquitylation is sufficient to drive these processes.
Protocol: Ligand-Independent Endocytosis of Membrane Proteins
Construct Design:
- Fuse the extracellular and transmembrane domains of a membrane protein (e.g., EGFR, CD4) to the CUbo tag
- Include an extracellular epitope tag for antibody detection
- Co-express with the NUbo tag fused to the K63-specific E3 (Pib1-Ubc13·Mms2)
Cell Surface Labeling:
- Plate transfected cells on glass coverslips
- Chill cells to 4°C to arrest membrane trafficking
- Label surface proteins with fluorescent antibodies (e.g., anti-HA Alexa Fluor 488) for 30 minutes on ice
Internalization Assay:
- Add rapamycin (500 nM) or vehicle control
- Quickly shift to 37°C to restart trafficking
- At time points (0, 5, 15, 30 minutes), fix cells with 4% PFA
Visualization and Quantification:
- Permeabilize fixed cells and stain with appropriate secondary antibodies if needed
- Image using confocal microscopy
- Quantify internalization by measuring the ratio of intracellular to surface fluorescence using image analysis software (e.g., ImageJ)
Expected Results: K63-polyubiquitylation is sufficient to induce rapid internalization of plasma membrane proteins, with significant internalization observed within 15-30 minutes of rapamycin addition [64]. This approach can determine whether K63 ubiquitylation directly drives endocytosis of a protein of interest, independent of other activation signals.
Implementation Across Model Systems
Validation in Budding Yeast
The Ubiquiton system was successfully implemented in Saccharomyces cerevisiae, demonstrating its applicability in genetically tractable model organisms.
Protocol: Yeast Transformation and Phenotypic Analysis
Strain Engineering:
- Integrate CUbo-tagged POI under endogenous promoter
- Express NUbo-E3 fusions from constitutive or inducible promoters
- Use standard lithium acetate transformation protocol
Rapamycin Treatment:
- Grow yeast to mid-log phase (OD600 = 0.5-0.8)
- Add rapamycin (1-10 nM final concentration) from fresh stock
- Monitor growth and phenotype over 6-24 hours
Protein Analysis:
- Collect samples at time points for protein extraction
- Use trichloroacetic acid (TCA) precipitation for protein preparation
- Analyze by Western blot with anti-ubiquitin or protein-specific antibodies
Adaptation to Mammalian Cell Systems
The system has been extensively validated in human cell lines, showing broad applicability across protein classes.
Key Considerations for Mammalian Cells:
- Use inducible expression systems to minimize potential toxicity
- Titrate rapamycin concentration (typically 100-500 nM)
- Include controls for rapamycin-induced signaling effects
- Consider cell type-specific variations in ubiquitin machinery
Table: Comparison of Ubiquiton Performance Across Model Systems
| Parameter | Budding Yeast | Mammalian Cells |
|---|---|---|
| Optimal Rapamycin Concentration | 1-10 nM | 100-500 nM |
| Degradation Half-life (K48) | ~5-15 minutes | ~5-30 minutes |
| Key Applications | Chromatin biology, DNA repair, basic proteostasis | Signaling pathways, membrane trafficking, disease modeling |
| Throughput Potential | High (genetic screens) | Moderate (arrayed formats) |
The Scientist's Toolkit: Essential Research Reagents
Successful implementation of the Ubiquiton system requires several key reagents and controls. The table below details essential components and their functions.
Table: Essential Research Reagents for Ubiquiton Studies
| Reagent/Category | Specific Examples | Function/Application | Considerations |
|---|---|---|---|
| Linkage-Specific E3s | HOIP(M1), Cue1-Ubc7(K48), Pib1-Ubc13·Mms2(K63) | Catalyze defined ubiquitin linkages | Specific E2 co-expression required |
| Detection Reagents | Linkage-specific antibodies, TUBEs [66], Mass spectrometry | Verify chain type and specificity | Antibody validation essential |
| DUB Inhibitors | PR-619 (pan-DUB inhibitor) [67] | Stabilize ubiquitin conjugates | Can have off-target effects |
| Rapamycin Analogs | Standard rapamycin, Shield-1 (alternative) | Induce dimerization | Dose optimization required |
| Control Plasmids | Catalytically dead E3 mutants (C91S OTUB1) [67] | Specificity controls | Essential for mechanistic studies |
| Model System Tools | Yeast genetic tools, CRISPR/Cas9 knockouts (RNF19A/B) [68] | System validation and genetic analysis | Cell line-specific optimization |
Critical Experimental Design Considerations
System Optimization and Validation
Proper Controls: Always include:
- Catalytically dead E3 mutants (e.g., OTUB1 C91S) to confirm enzymatic specificity [67]
- Rapamycin-only treatments to exclude dimerizer effects
- Untagged POI to confirm tagging-dependent effects
- Linkage-specific E3 mutants to verify linkage dependency
Validation Experiments:
- Verify Ubiquitin Chain Linkage: Use linkage-specific antibodies [69] or Tandem Ubiquitin Binding Entities (TUBEs) [66] to confirm intended chain formation
- Assess Background Activity: Monitor system activity without rapamycin induction
- Determine Kinetics: Establish time and concentration curves for rapamycin induction
- Evaluate Off-target Effects: Use proteomic approaches to assess system specificity
Advanced Applications and Integration
The following diagram illustrates how Ubiquiton integrates with modern ubiquitin research tools and potential therapeutic applications:
The Ubiquiton system represents a versatile platform that integrates with modern ubiquitin research tools including UbiREAD for decoding ubiquitin chains [65], PROTACs for targeted protein degradation [70], DUBTACs for protein stabilization [67], and activity-based probes for deubiquitinase profiling [71]. This integration enables comprehensive investigation of ubiquitin-dependent processes across model systems and protein classes.
The Ubiquiton system provides researchers with an unprecedented ability to control linkage-specific polyubiquitylation in living cells, offering remarkable versatility across model systems—from budding yeast to mammalian cells—and diverse protein classes including soluble cytoplasmic proteins, nuclear proteins, chromatin-associated factors, and integral membrane proteins [64]. By enabling precise manipulation of the ubiquitin code, this technology accelerates the dissection of ubiquitin-dependent processes in proteostasis, signaling, trafficking, and DNA repair. The protocols and applications detailed in this document provide a foundation for researchers to implement this powerful tool in their investigation of ubiquitin biology and its roles in health and disease.
Conclusion
The Ubiquiton system marks a significant leap forward in our ability to interrogate the ubiquitin code with precision. By providing researchers with an inducible, linkage-specific tool, it overcomes the major limitations of traditional, non-specific approaches to studying ubiquitination. The validated applications in controlling protein stability, localization, and signaling open new avenues for basic research into cellular homeostasis, cancer biology, and neurodegenerative disorders. For drug discovery, Ubiquiton serves as both a powerful validation tool for targeting ubiquitin pathways and a platform that could inspire novel therapeutic strategies, such as the next generation of targeted degraders. Future developments will likely focus on expanding the repertoire of E3 ligases and ubiquitin linkages within the system, further solidifying its role as an indispensable resource for decoding ubiquitin signaling and advancing human health.
References
- 1. Molecular choreography of E1 enzymes in ubiquitin-like ... [https://pubmed.ncbi.nlm.nih.gov/40570956/]
- 2. Quantifying Ubiquitin Signaling - PMC - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC4441763/]
- 3. Structure-guided engineering enables E3 ligase-free and ... [https://www.nature.com/articles/s41467-024-45635-y]
- 4. Ubiquitination-mediated molecular pathway alterations in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9520991/]
- 5. A cascading activity-based probe sequentially targets E1 ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5108872/]
- 6. Structural insights into the biochemical mechanism of ... [https://www.sciencedirect.com/science/article/abs/pii/S0969212624005379]
- 7. Ubiquiton-An inducible, linkage-specific polyubiquitylation ... [https://pubmed.ncbi.nlm.nih.gov/38103558/]
- 8. An Inducible, Linkage-Specific Polyubiquitylation Tool [https://papers.ssrn.com/sol3/papers.cfm?abstract_id=4421571]
- 9. Ubiquiton-An inducible, linkage-specific polyubiquitylation ... [https://www.merckmillipore.com/SR/en/tech-docs/paper/1640831]
- 10. Breaking the K48-Chain: Linking Ubiquitin Beyond Protein ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11730971/]
- 11. Current methodologies in protein ubiquitination ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9373315/]
- 12. Combinatorial ubiquitin code degrades deubiquitylation- ... [https://www.nature.com/articles/s41467-025-57873-9]
- 13. Unraveling chain specific ubiquitination in cells using ... [https://www.nature.com/articles/s41598-025-07242-9]
- 14. The Role of Protein Ubiquitination in the Onset and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12248740/]
- 15. Ubiquiton—An inducible, linkage-specific ... [https://www.sciencedirect.com/science/article/pii/S1097276523009619]
- 16. Current methodologies in protein ubiquitination characterization [https://cellandbioscience.biomedcentral.com/articles/10.1186/s13578-022-00870-y]
- 17. Ubiquiton – an inducible, linkage-specific polyubiquitylation tool [https://www.omicsdi.org/dataset/pride/PXD045662]
- 18. Protocol for In Vitro Ubiquitin Conjugation Reaction [https://www.rndsystems.com/resources/protocols/vitro-ubiquitin-conjugation-reaction]
- 19. A Data Set of Human Endogenous Protein Ubiquitination ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3098580/]
- 20. Quantitative Proteomic Atlas of Ubiquitination and Acetylation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4560960/]
- 21. Chemical approaches to explore ubiquitin-like proteins [https://pubs.rsc.org/en/content/articlehtml/2025/cb/d4cb00220b]
- 22. BioE3 identifies specific substrates of ubiquitin E3 ligases [https://www.nature.com/articles/s41467-023-43326-8]
- 23. An engineered platform for reconstituting functional ... [https://www.sciencedirect.com/science/article/pii/S1674205222001939]
- 24. Structural visualization of HECT-type E3 ligase Ufd4 ... [https://www.nature.com/articles/s41467-025-59569-6]
- 25. Origin and Function of Ubiquitin-like Protein Conjugation - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC2819001/]
- 26. Publication: Ubiquiton - An inducible , linkage - specific ... [https://www.imb.de/publication/ubiquiton-an-inducible-linkage-specific-polyubiquitylation-tool]
- 27. Decoding polyubiquitin regulation of KV7. 1 (KCNQ1) ... [https://www.nature.com/articles/s41467-025-60893-0]
- 28. Cypin regulates K63-linked polyubiquitination to shape ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12248297/]
- 29. Site-directed multivalent conjugation of antibodies to ... [https://www.nature.com/articles/s41551-024-01342-z]
- 30. Conjugation kits | abcam [https://www.abcam.com/en-us/products/conjugation-kits]
- 31. AGLink® Site-specific conjugation kit [https://www.acrobiosystems.com/category/kits-assay-reagents/adc-conjugationkit?srsltid=AfmBOopr6Q1RjC4gZGV6Sa1tkISaeTmqzivOqee6nsxUZSgASHNoQlbG]
- 32. Ubiquitin Ligase Substrate Identification through ... [https://www.sciencedirect.com/science/article/pii/S002192582087207X]
- 33. Next-Generation Protein Degradation - 2025 Archive [https://www.chi-peptalk.com/25/protacs-protein-degradation]
- 34. Ubiquitination and deubiquitination in cancer [https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-024-02046-3]
- 35. A family of E3 ligases extend K11 polyubiquitin on sites ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12132378/]
- 36. Structural basis of ubiquitin ligase Nedd4-2 autoinhibition ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12106849/]
- 37. Comparative CRISPRi screens reveal a human stem cell ... [https://www.nature.com/articles/s41594-025-01616-3]
- 38. Target screening and optimization of candidate ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12098522/]
- 39. Protein monoubiquitylation: targets and diverse functions - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4744734/]
- 40. E3 ubiquitin ligases: styles, structures and functions - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC8607428/]
- 41. substrate recognition and catalysis by ubiquitin ligases [https://www.sciencedirect.com/science/article/pii/S0962892400018341]
- 42. Structural mechanisms of autoinhibition and substrate ... [https://www.nature.com/articles/s41594-023-01203-4]
- 43. A high sensitivity assay of UBE3A ubiquitin ligase activity [https://pubmed.ncbi.nlm.nih.gov/39933617/]
- 44. Chemical Tools for Probing the Ub/Ubl Conjugation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11727022/]
- 45. Conceptualizing Experimental Controls Using the Potential ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12609828/]
- 46. Data Interpretation in Research | Overview, Software & ... [https://atlasti.com/guides/qualitative-research-guide-part-2/data-interpretation]
- 47. Module 2: Research Design - Section 2 | ORI [https://ori.hhs.gov/module-2-research-design-section-2]
- 48. Quantitative Proteomics Reveals the Function of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2668214/]
- 49. Ubiquitin-dependent endocytosis, trafficking and turnover of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3279607/]
- 50. Visualizing the functional architecture of the endocytic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4357291/]
- 51. Emerging functions of branched ubiquitin chains [https://www.nature.com/articles/s41421-020-00237-y]
- 52. UbiREAD deciphers proteasomal degradation code of ... [https://www.sciencedirect.com/science/article/pii/S1097276525001522]
- 53. Design and development of PROTACs: A new paradigm in ... [https://www.sciencedirect.com/science/article/pii/S2590098625000181]
- 54. Molecular Design of Novel Protein-Degrading ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12196083/]
- 55. Systematic comparison and base-editing-mediated ... [https://www.nature.com/articles/s41467-025-61848-1]
- 56. Controllable targeted protein degradation as a promising ... [https://www.sciencedirect.com/science/article/abs/pii/S0012160625002647]
- 57. N-Degron-Based PROTAC Targeting PLK1: A Potential ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12389170/]
- 58. Ubiquitinâ€Independent Degradation: An Emerging PROTAC ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11755708/]
- 59. PROTAC Degraders in Clinical Trails: 2025 Update [https://www.biochempeg.com/article/434.html]
- 60. Mechanisms of Generating Polyubiquitin Chains ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4197637/]
- 61. Ubiquiton-An inducible, linkage-specific polyubiquitylation ... [https://www.addgene.org/browse/article/28243691/]
- 62. Ubiquiton-An inducible, linkage-specific polyubiquitylation ... [https://www.merckmillipore.com/CI/fr/tech-docs/paper/1640831]
- 63. Mechanisms underlying linear ubiquitination and implications ... [https://biosignaling.biomedcentral.com/articles/10.1186/s12964-023-01239-5]
- 64. Ubiquiton—An inducible, linkage-specific ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10804999/]
- 65. Cracking the Code of Protein Degradation [https://www.mpg.de/24380519/cracking-the-code-of-protein-degradation]
- 66. HTS Screening of Poly-Ubiquitin Linkage-Specific ... [https://lifesensors.com/hts-screening-of-poly-ubiquitin-linkage-specific-molecular-glues-and-protein-degraders/]
- 67. Programmable protein stabilization with language model ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11997201/]
- 68. Highly specific intracellular ubiquitination of a small molecule [https://www.nature.com/articles/s41589-025-02011-1]
- 69. The Molecular Toolbox for Linkage Type-Specific Analysis ... [https://pubmed.ncbi.nlm.nih.gov/40192223/]
- 70. Focus on ubiquitylation and protein degradation [https://www.nature.com/articles/s41594-025-01672-9]
- 71. Chemical tools to define and manipulate interferon ... [https://www.nature.com/articles/s41467-025-56336-5]